Directed evolution of Methanococcus jannaschii citramalate synthase for biosynthesis of 1-propanol and 1-butanol by Escherichia coli
- PMID: 18952866
- PMCID: PMC2607174
- DOI: 10.1128/AEM.02046-08
Directed evolution of Methanococcus jannaschii citramalate synthase for biosynthesis of 1-propanol and 1-butanol by Escherichia coli
Abstract
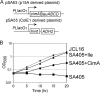
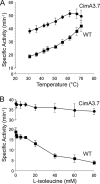
Biofuels synthesized from renewable resources are of increasing interest because of global energy and environmental problems. We have previously demonstrated production of higher alcohols from Escherichia coli using a 2-keto acid-based pathway. Here, we took advantage of the growth phenotype associated with 2-keto acid deficiency to construct a hyperproducer of 1-propanol and 1-butanol by evolving citramalate synthase (CimA) from Methanococcus jannaschii. This new pathway, which directly converts pyruvate to 2-ketobutyrate, bypasses threonine biosynthesis and represents the shortest keto acid-mediated pathway for producing 1-propanol and 1-butanol from glucose. Directed evolution of CimA enhanced the specific activity over a wide temperature range (30 to 70 degrees C). The best CimA variant was found to be insensitive to feedback inhibition by isoleucine in addition to the improved activity. This CimA variant enabled 9- and 22-fold higher production levels of 1-propanol and 1-butanol, respectively, compared to the strain expressing the wild-type CimA. This work demonstrates (i) the first production of 1-propanol and 1-butanol using the citramalate pathway and (ii) the benefit of the 2-keto acid pathway that enables a growth-based evolutionary strategy to improve the production of non-growth-related products.
Figures





References
-
- Atsumi, S., T. Hanai, and J. C. Liao. 2008. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86-89. - PubMed
-
- Cadwell, R. C., and G. F. Joyce. 1994. Mutagenic PCR. PCR Methods Appl. 3:S136-S140. - PubMed
-
- Eoyang, L., and P. M. Silverman. 1988. Purification and assays of acetolactate synthase I from Escherichia coli K12. Methods Enzymol. 166:435-445. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

