Positional distribution of human transcription factor binding sites
- PMID: 18953043
- PMCID: PMC2588498
- DOI: 10.1093/nar/gkn752
Positional distribution of human transcription factor binding sites
Abstract
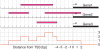
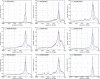
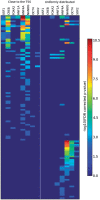
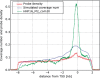
We developed a method for estimating the positional distribution of transcription factor (TF) binding sites using ChIP-chip data, and applied it to recently published experiments on binding sites of nine TFs: OCT4, SOX2, NANOG, HNF1A, HNF4A, HNF6, FOXA2, USF1 and CREB1. The data were obtained from a genome-wide coverage of promoter regions from 8-kb upstream of the transcription start site (TSS) to 2-kb downstream. The number of target genes of each TF ranges from few hundred to several thousand. We found that for each of the nine TFs the estimated binding site distribution is closely approximated by a mixture of two components: a narrow peak, localized within 300-bp upstream of the TSS, and a distribution of almost uniform density within the tested region. Using Gene Ontology (GO) and Enrichment analysis, we were able to associate (for each of the TFs studied) the target genes of both types of binding with known biological processes. Most GO terms were enriched either among the proximal targets or among those with a uniform distribution of binding sites. For example, the three stemness-related TFs have several hundred target genes that belong to 'development' and 'morphogenesis' whose binding sites belong to the uniform distribution.
Figures



References
-
- Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat. Rev. Genet. 2006;7:55–65. - PubMed
-
- Rodriguez BA, Huang TH. Tilling the chromatin landscape: emerging methods for the discovery and profiling of protein-DNA interactions. Biochem. Cell. Biol. 2005;83:525–534. - PubMed
-
- Hertzberg L, Zuk O, Getz G, Domany E. Finding motifs in promoter regions. J. Comput. Biol. 2005;12:314–330. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

