Adaptation to cell culture induces functional differences in measles virus proteins
- PMID: 18954437
- PMCID: PMC2582235
- DOI: 10.1186/1743-422X-5-129
Adaptation to cell culture induces functional differences in measles virus proteins
Abstract
Background: Live, attenuated measles virus (MeV) vaccine strains were generated by adaptation to cell culture. The genetic basis for the attenuation of the vaccine strains is unknown. We previously reported that adaptation of a pathogenic, wild-type MeV to Vero cells or primary chicken embryo fibroblasts (CEFs) resulted in a loss of pathogenicity in rhesus macaques. The CEF-adapted virus (D-CEF) contained single amino acid changes in the C and matrix (M) proteins and two substitutions in the shared amino terminal domain of the phosphoprotein (P) and V protein. The Vero-adapted virus (D-VI) had a mutation in the cytoplasmic tail of the hemagglutinin (H) protein.
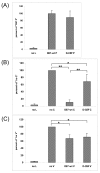
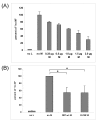
Results: In vitro assays were used to test the functions of the wild-type and mutant proteins. The substitution in the C protein of D-CEF decreased its ability to inhibit mini-genome replication, while the wild-type and mutant M proteins inhibited replication to the same extent. The substitution in the cytoplasmic tail of the D-VI H protein resulted in reduced fusion in a quantitative fusion assay. Co-expression of M proteins with wild-type fusion and H proteins decreased fusion activity, but the mutation in the M protein of D-CEF did not affect this function. Both mutations in the P and V proteins of D-CEF reduced the ability of these proteins to inhibit type I and II interferon signaling.
Conclusion: Adaptation of a wild-type MeV to cell culture selected for genetic changes that caused measurable functional differences in viral proteins.
Figures


References
-
- Ikic D, Juzbasic M, Beck M, Hrabar A, Cimbur-Schreiber T. Attenuation and characterisation of Edmonston-Zagreb measles virus. Ann Immunol Hung. 1972;16:175–181. - PubMed
-
- Sasaki K. Studies on the modification of the live AIK measles vaccine. I. Adaptation of the further attenuated AIK measles virus (the strain AIK-L33) to chick embryo cells. Kitasato Arch Exp Med. 1974;47:1–12. - PubMed
-
- Schwarz AJ. Immunization against Measles: Development and Evaluation of a Highly Attenuated Live Measles Vaccine. Ann Paediatr. 1964;202:241–252. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

