Residue 17 of sauvagine cross-links to the first transmembrane domain of corticotropin-releasing factor receptor 1 (CRFR1)
- PMID: 18955489
- PMCID: PMC2602896
- DOI: 10.1074/jbc.M806351200
Residue 17 of sauvagine cross-links to the first transmembrane domain of corticotropin-releasing factor receptor 1 (CRFR1)
Abstract
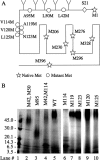
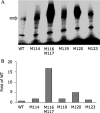
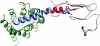
Corticotropin-releasing factor receptor 1 (CRFR1) mediates the physiological actions of corticotropin-releasing factor in the anterior pituitary gland and the central nervous system. Using chemical cross-linking we have previously reported that residue 16 of sauvagine (SVG) is in a close proximity to the second extracellular loop of CRFR1. Here we introduced p-benzoylphenylalanine (Bpa) at position 17 of a sauvagine analog, [Tyr0, Gln1, Bpa17]SVG, to covalently label CRFR1 and characterize the cross-linking site. Using a combination of receptor mutagenesis, peptide mapping, and N-terminal sequencing, we identified His117 within the first transmembrane domain (TM1) of CRFR1 as the cross-linking site for Bpa17 of 125I-[Tyr0, Gln1, Bpa17]SVG. These data indicate that, within the SVG-CRFR1 complex, residue 17 of the ligand lies within a 9 angstroms distance from residue 117 of the TM1 of CRFR1. The molecular proximity between residue 17 of the ligand and TM1 of CRFR1 described here and between residue 16 of the ligand and the CRFR1 second extracellular loop described previously provides useful molecular constraints for modeling ligand-receptor interaction in mammalian cells expressing CRFR1.
Figures
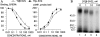


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

