Endothelial adhesion receptors are recruited to adherent leukocytes by inclusion in preformed tetraspanin nanoplatforms
- PMID: 18955551
- PMCID: PMC2575792
- DOI: 10.1083/jcb.200805076
Endothelial adhesion receptors are recruited to adherent leukocytes by inclusion in preformed tetraspanin nanoplatforms
Abstract
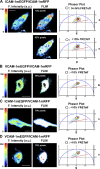
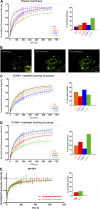
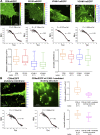
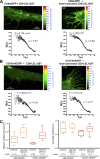
VCAM-1 and ICAM-1, receptors for leukocyte integrins, are recruited to cell-cell contact sites on the apical membrane of activated endothelial cells. In this study, we show that this recruitment is independent of ligand engagement, actin cytoskeleton anchorage, and heterodimer formation. Instead, VCAM-1 and ICAM-1 are recruited by inclusion within specialized preformed tetraspanin-enriched microdomains, which act as endothelial adhesive platforms (EAPs). Using advanced analytical fluorescence techniques, we have characterized the diffusion properties at the single-molecule level, nanoscale organization, and specific intradomain molecular interactions of EAPs in living primary endothelial cells. This study provides compelling evidence for the existence of EAPs as physical entities at the plasma membrane, distinct from lipid rafts. Scanning electron microscopy of immunogold-labeled samples treated with a specific tetraspanin-blocking peptide identify nanoclustering of VCAM-1 and ICAM-1 within EAPs as a novel mechanism for supramolecular organization that regulates the leukocyte integrin-binding capacity of both endothelial receptors during extravasation.
Figures



Comment in
-
Dances with leukocytes: how tetraspanin-enriched microdomains assemble to form endothelial adhesive platforms.J Cell Biol. 2008 Nov 3;183(3):375-6. doi: 10.1083/jcb.200809173. J Cell Biol. 2008. PMID: 18981226 Free PMC article.
References
-
- Anderson, R.G., and K. Jacobson. 2002. A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science. 296:1821–1825. - PubMed
-
- Barreiro, O., M. Yanez-Mo, J.M. Serrador, M.C. Montoya, M. Vicente-Manzanares, R. Tejedor, H. Furthmayr, and F. Sanchez-Madrid. 2002. Dynamic interaction of VCAM-1 and ICAM-1 with moesin and ezrin in a novel endothelial docking structure for adherent leukocytes. J. Cell Biol. 157:1233–1245. - PMC - PubMed
-
- Barreiro, O., M. Yanez-Mo, M. Sala-Valdes, M.D. Gutierrez-Lopez, S. Ovalle, A. Higginbottom, P.N. Monk, C. Cabanas, and F. Sanchez-Madrid. 2005. Endothelial tetraspanin microdomains regulate leukocyte firm adhesion during extravasation. Blood. 105:2852–2861. - PubMed
-
- Berditchevski, F. 2001. Complexes of tetraspanins with integrins: more than meets the eye. J. Cell Sci. 114:4143–4151. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

