Analysis of histones in Xenopus laevis. I. A distinct index of enriched variants and modifications exists in each cell type and is remodeled during developmental transitions
- PMID: 18957438
- PMCID: PMC2613616
- DOI: 10.1074/jbc.M807273200
Analysis of histones in Xenopus laevis. I. A distinct index of enriched variants and modifications exists in each cell type and is remodeled during developmental transitions
Abstract
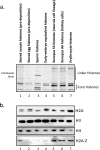
Histone proteins contain epigenetic information that is encoded both in the relative abundance of core histones and variants and particularly in the post-translational modification of these proteins. We determined the presence of such variants and covalent modifications in seven tissue types of the anuran Xenopus laevis, including oocyte, egg, sperm, early embryo equivalent (pronuclei incubated in egg extract), S3 neurula cells, A6 kidney cells, and erythrocytes. We first developed a new robust method for isolating the stored, predeposition histones from oocytes and eggs via chromatography on heparin-Sepharose, whereas we isolated chromatinized histones via conventional acid extraction. We identified two previously unknown H1 isoforms (H1fx and H1B.Sp) present on sperm chromatin. We immunoblotted this global collection of histones with many specific post-translational modification antibodies, including antibodies against methylated histone H3 on Lys(4), Lys(9), Lys(27), Lys(79), Arg(2), Arg(17), and Arg(26); methylated histone H4 on Lys(20); methylated H2A and H4 on Arg(3); acetylated H4 on Lys(5), Lys(8), Lys(12), and Lys(16) and H3 on Lys(9) and Lys(14); and phosphorylated H3 on Ser(10) and H2A/H4 on Ser(1). Furthermore, we subjected a subset of these histones to two-dimensional gel analysis and subsequent immunoblotting and mass spectrometry to determine the global remodeling of histone modifications that occurs as development proceeds. Overall, our observations suggest that each metazoan cell type may have a unique histone modification signature correlated with its differentiation status.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

