Quantitative expression of the Candida albicans secreted aspartyl proteinase gene family in human oral and vaginal candidiasis
- PMID: 18957581
- PMCID: PMC2722715
- DOI: 10.1099/mic.0.2008/022293-0
Quantitative expression of the Candida albicans secreted aspartyl proteinase gene family in human oral and vaginal candidiasis
Abstract
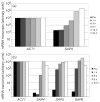
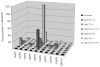
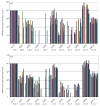
A quantitative real-time RT-PCR system was established to identify which secreted aspartyl proteinase (SAP) genes are most highly expressed and potentially contribute to Candida albicans infection of human epithelium in vitro and in vivo. C. albicans SC5314 SAP1-10 gene expression was monitored in organotypic reconstituted human epithelium (RHE) models, monolayers of oral epithelial cells, and patients with oral (n=17) or vaginal (n=17) candidiasis. SAP gene expression was also analysed in Deltasap1-3, Deltasap4-6, Deltaefg1 and Deltaefg1/cph1 mutants to determine whether compensatory SAP gene regulation occurs in the absence of distinct proteinase gene subfamilies. In monolayers, RHE models and patient samples SAP9 was consistently the most highly expressed gene in wild-type cells. SAP5 was the only gene significantly upregulated as infection progressed in both RHE models and was also highly expressed in patient samples. Interestingly, the SAP4-6 subfamily was generally more highly expressed in oral monolayers than in RHE models. SAP1 and SAP2 expression was largely unchanged in all model systems, and SAP3, SAP7 and SAP8 were expressed at low levels throughout. In Deltasap1-3, expression was compensated for by increased expression of SAP5, and in Deltasap4-6, expression was compensated for by SAP2: both were observed only in the oral RHE. Both Deltasap1-3 and Deltasap4-6 mutants caused RHE tissue damage comparable to the wild-type. However, addition of pepstatin A reduced tissue damage, indicating a role for the Sap family as a whole in inducing epithelial damage. With the hypha-deficient mutants, RHE tissue damage was significantly reduced in both Deltaefg1/cph1 and Deltaefg1, but SAP5 expression was only dramatically reduced in Deltaefg1/cph1 despite the absence of hyphal growth in both mutants. This indicates that hypha formation is the predominant cause of tissue damage, and that SAP5 expression can be hypha-independent and is not solely controlled by the Efg1 pathway but also by the Cph1 pathway. This is believed to be the first study to fully quantify SAP gene expression levels during human mucosal infections; the results suggest that SAP5 and SAP9 are the most highly expressed proteinase genes in vivo. However, the overall contribution of the Sap1-3 and Sap4-6 subfamilies individually in inducing epithelial damage in the RHE models appears to be low.
Figures
References
-
- Albrecht A, Felk A, Pichova I, Schaller M, de Groot P, MacCallum D, Odds FC, Schäfer W, Klis F, et al. Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J Biol Chem. 2006;281:688–694. - PubMed
-
- Felk A, Kretschmar M, Albrecht A, Schaller M, Beinhauer S, Nichterlein T, Sanglard D, Korting HC, Schafer W, Hube B. Candida albicans hyphal formation and the expression of the Efg1-regulated proteinases Sap4 to Sap6 are required for the invasion of parenchymal organs. Infect Immun. 2002;70:3689–3700. - PMC - PubMed
-
- Frank CF, Hostetter MK. Cleavage of E-cadherin: a mechanism for disruption of the intestinal epithelial barrier by Candida albicans. Transl Res. 2007;149:211–222. - PubMed
-
- Gillum AM, Tsay EY, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–182. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

