Rapid high resolution single nucleotide polymorphism-comparative genome hybridization mapping in Caenorhabditis elegans
- PMID: 18957702
- PMCID: PMC2621182
- DOI: 10.1534/genetics.108.096487
Rapid high resolution single nucleotide polymorphism-comparative genome hybridization mapping in Caenorhabditis elegans
Abstract
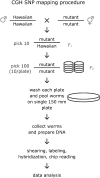
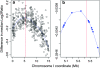
We have developed a significantly improved and simplified method for high-resolution mapping of phenotypic traits in Caenorhabditis elegans using a combination of single nucleotide polymorphisms (SNPs) and oligo array comparative genome hybridization (array CGH). We designed a custom oligonucleotide array using a subset of confirmed SNPs between the canonical wild-type Bristol strain N2 and the Hawaiian isolate CB4856, populated with densely overlapping 50-mer probes corresponding to both N2 and CB4856 SNP sequences. Using this method a mutation can be mapped to a resolution of approximately 200 kb in a single genetic cross. Six mutations representing each of the C. elegans chromosomes were detected unambiguously and at high resolution using genomic DNA from populations derived from as few as 100 homozygous mutant segregants of mutant N2/CB4856 heterozygotes. Our method completely dispenses with the PCR, restriction digest, and gel analysis of standard SNP mapping and should be easy to extend to any organism with interbreeding strains. This method will be particularly powerful when applied to difficult or hard-to-map low-penetrance phenotypes. It should also be possible to map polygenic traits using this method.
Figures


References
-
- Chen, D., A. Ahlford, F. Schnorrer, I. Kalchhauser, M. Fellner et al., 2008. High-resolution, high-throughput SNP mapping in Drosophila melanogaster. Nat. Methods 5 323–329. - PubMed
-
- Hillier, L.W., G. T. Marth, A. R. Quinlin, D. Dooling, G. Fewell et al., 2008. Whole-genome sequencing and variant discovery in C. elegans. Nat. Methods 5 183–188. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

