Patterns of hybrid loss of imprinting reveal tissue- and cluster-specific regulation
- PMID: 18958286
- PMCID: PMC2570336
- DOI: 10.1371/journal.pone.0003572
Patterns of hybrid loss of imprinting reveal tissue- and cluster-specific regulation
Abstract
Background: Crosses between natural populations of two species of deer mice, Peromyscus maniculatus (BW), and P. polionotus (PO), produce parent-of-origin effects on growth and development. BW females mated to PO males (bwxpo) produce growth-retarded but otherwise healthy offspring. In contrast, PO females mated to BW males (POxBW) produce overgrown and severely defective offspring. The hybrid phenotypes are pronounced in the placenta and include POxBW conceptuses which lack embryonic structures. Evidence to date links variation in control of genomic imprinting with the hybrid defects, particularly in the POxBW offspring. Establishment of genomic imprinting is typically mediated by gametic DNA methylation at sites known as gDMRs. However, imprinted gene clusters vary in their regulation by gDMR sequences.
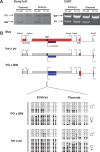
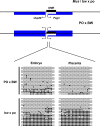
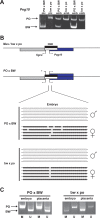
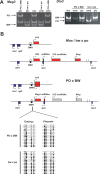
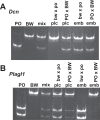
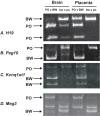
Methodology/principal findings: Here we further assess imprinted gene expression and DNA methylation at different cluster types in order to discern patterns. These data reveal POxBW misexpression at the Kcnq1ot1 and Peg3 clusters, both of which lose ICR methylation in placental tissues. In contrast, some embryonic transcripts (Peg10, Kcnq1ot1) reactivated the silenced allele with little or no loss of DNA methylation. Hybrid brains also display different patterns of imprinting perturbations. Several cluster pairs thought to use analogous regulatory mechanisms are differentially affected in the hybrids.
Conclusions/significance: These data reinforce the hypothesis that placental and somatic gene regulation differs significantly, as does that between imprinted gene clusters and between species. That such epigenetic regulatory variation exists in recently diverged species suggests a role in reproductive isolation, and that this variation is likely to be adaptive.
Conflict of interest statement
Figures
References
-
- Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. - PubMed
-
- Tanaka K, Shiota G, Meguro M, Mitsuya K, Oshimura M, et al. Loss of imprinting of long QT intronic transcript 1 in colorectal cancer. Oncology. 2001;60:268–273. - PubMed
-
- Fisher RA, Hodges MD. Genomic imprinting in gestational trophoblastic disease–a review. Placenta. 2003;(Suppl A):S111–1118. - PubMed
-
- Delaval K, Wagschal A, Feil R. Epigenetic deregulation of imprinting in congenital diseases of aberrant growth. Bioessays. 2006;28:453–459. - PubMed
-
- Mackay DJ, Hahnemann JM, Boonen SE, Poerksen S, Bunyan DJ, et al. Epimutation of the TNDM locus and the Beckwith-Wiedemann syndrome centromeric locus in individuals with transient neonatal diabetes mellitus. Hum Genet. 2006;119:179–184. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

