Federated access to heterogeneous information resources in the Neuroscience Information Framework (NIF)
- PMID: 18958629
- PMCID: PMC2689790
- DOI: 10.1007/s12021-008-9033-y
Federated access to heterogeneous information resources in the Neuroscience Information Framework (NIF)
Abstract
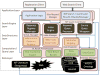
The overarching goal of the NIF (Neuroscience Information Framework) project is to be a one-stop-shop for Neuroscience. This paper provides a technical overview of how the system is designed. The technical goal of the first version of the NIF system was to develop an information system that a neuroscientist can use to locate relevant information from a wide variety of information sources by simple keyword queries. Although the user would provide only keywords to retrieve information, the NIF system is designed to treat them as concepts whose meanings are interpreted by the system. Thus, a search for term should find a record containing synonyms of the term. The system is targeted to find information from web pages, publications, databases, web sites built upon databases, XML documents and any other modality in which such information may be published. We have designed a system to achieve this functionality. A central element in the system is an ontology called NIFSTD (for NIF Standard) constructed by amalgamating a number of known and newly developed ontologies. NIFSTD is used by our ontology management module, called OntoQuest to perform ontology-based search over data sources. The NIF architecture currently provides three different mechanisms for searching heterogeneous data sources including relational databases, web sites, XML documents and full text of publications. Version 1.0 of the NIF system is currently in beta test and may be accessed through http://nif.nih.gov.
Figures




References
-
- Astakhov V, Gupta A, Grethe JS, Ross E, Little D, Yilmaz A, Qian X, Santini S, Martone ME, Ellisman M. Semantically based data integration environment for biomedical research. Proc. 19th IEEE Symposium on Computer-Based Medical Systems; IEEE Computer Society, Washington, DC, USA. 2006. pp. 171–176.
-
- Chen L, Gupta A, Kurul ME. Stack-based algorithms for pattern matching on dags. Proc. 31st Int. Conf. on Very Large Databases (VLDB); Stockholm. 2005. pp. 493–504.
-
- Chen L, Martone ME, Gupta A, Fong L, Wong-Barnum M. Ontoquest: Exploring ontological data made easy. Proc. 31st Int. Conf. on Very Large Databases (VLDB); 2006. pp. 1183–1186.
-
- Franklin MJ, Halevy AY, Maier D. From databases to dataspaces: a new abstraction for information management. SIGMOD Record. 2005;34(4):27–33.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

