minet: A R/Bioconductor package for inferring large transcriptional networks using mutual information
- PMID: 18959772
- PMCID: PMC2630331
- DOI: 10.1186/1471-2105-9-461
minet: A R/Bioconductor package for inferring large transcriptional networks using mutual information
Abstract
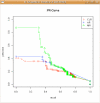
Results: This paper presents the R/Bioconductor package minet (version 1.1.6) which provides a set of functions to infer mutual information networks from a dataset. Once fed with a microarray dataset, the package returns a network where nodes denote genes, edges model statistical dependencies between genes and the weight of an edge quantifies the statistical evidence of a specific (e.g transcriptional) gene-to-gene interaction. Four different entropy estimators are made available in the package minet (empirical, Miller-Madow, Schurmann-Grassberger and shrink) as well as four different inference methods, namely relevance networks, ARACNE, CLR and MRNET. Also, the package integrates accuracy assessment tools, like F-scores, PR-curves and ROC-curves in order to compare the inferred network with a reference one.
Conclusion: The package minet provides a series of tools for inferring transcriptional networks from microarray data. It is freely available from the Comprehensive R Archive Network (CRAN) as well as from the Bioconductor website.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials