Changes in protein expression during honey bee larval development
- PMID: 18959778
- PMCID: PMC2760883
- DOI: 10.1186/gb-2008-9-10-r156
Changes in protein expression during honey bee larval development
Abstract
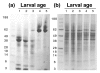
Background: The honey bee (Apis mellifera), besides its role in pollination and honey production, serves as a model for studying the biochemistry of development, metabolism, and immunity in a social organism. Here we use mass spectrometry-based quantitative proteomics to quantify nearly 800 proteins during the 5- to 6-day larval developmental stage, tracking their expression profiles.
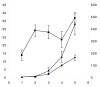
Results: We report that honey bee larval growth is marked by an age-correlated increase of protein transporters and receptors, as well as protein nutrient stores, while opposite trends in protein translation activity and turnover were observed. Levels of the immunity factors prophenoloxidase and apismin are positively correlated with development, while others surprisingly were not significantly age-regulated, suggesting a molecular explanation for why bees are susceptible to major age-associated bee bacterial infections such as American Foulbrood or fungal diseases such as chalkbrood. Previously unreported findings include the reduction of antioxidant and G proteins in aging larvae.
Conclusion: These data have allowed us to integrate disparate findings in previous studies to build a model of metabolism and maturity of the immune system during larval development. This publicly accessible resource for protein expression trends will help generate new hypotheses in the increasingly important field of honey bee research.
Figures



References
-
- Haldane JBS. Aristotle's account of bees' 'dances'. J Hellenic Studies. 1955;75:24–25. doi: 10.2307/629165. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

