Activation of pattern recognition receptor-mediated innate immunity inhibits the replication of hepatitis B virus in human hepatocyte-derived cells
- PMID: 18971270
- PMCID: PMC2612386
- DOI: 10.1128/JVI.02008-08
Activation of pattern recognition receptor-mediated innate immunity inhibits the replication of hepatitis B virus in human hepatocyte-derived cells
Abstract
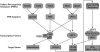
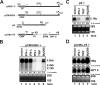
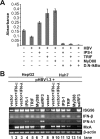
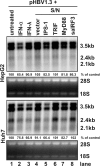
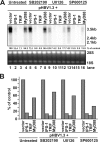
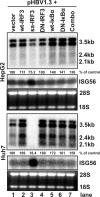
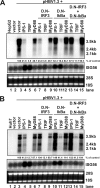
Recognition of virus infections by pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs), retinoic acid-inducible gene I (RIG-I), and melanoma differentiation associated gene 5 (MDA5), activates signaling pathways, leading to the induction of inflammatory cytokines that limit viral replication. To determine the effects of PRR-mediated innate immune response on hepatitis B virus (HBV) replication, a 1.3mer HBV genome was cotransfected into HepG2 or Huh7 cells with plasmid expressing TLR adaptors, myeloid differentiation primary response gene 88 (MyD88), and TIR-domain-containing adaptor-inducing beta interferon (TRIF), or RIG-I/MDA5 adaptor, interferon promoter stimulator 1 (IPS-1). The results showed that expressing each of the three adaptors dramatically reduced the levels of HBV mRNA and DNA in both HepG2 and Huh7 cells. However, HBV replication was not significantly affected by treatment of HBV genome-transfected cells with culture media harvested from cells transfected with each of the three adaptors, indicating that the adaptor-induced antiviral response was predominantly mediated by intracellular factors rather than by secreted cytokines. Analyses of involved signaling pathways revealed that activation of NF-kappaB is required for all three adaptors to elicit antiviral response in both HepG2 and Huh7 cells. However, activation of interferon regulatory factor 3 is only essential for induction of antiviral response by IPS-1 in Huh7 cells, but not in HepG2 cells. Furthermore, our results suggest that besides NF-kappaB, additional signaling pathway(s) are required for TRIF to induce a maximum antiviral response against HBV. Knowing the molecular mechanisms by which PRR-mediated innate defense responses control HBV infections could potentially lead to the development of novel therapeutics that evoke the host cellular innate antiviral response to control HBV infections.
Figures

References
-
- Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell 124783-801. - PubMed
-
- Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413732-738. - PubMed
-
- Ambros, V. 2004. The functions of animal microRNAs. Nature 431350-355. - PubMed
-
- Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116281-297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

