Genetic history of hepatitis C virus in East Asia
- PMID: 18971279
- PMCID: PMC2612398
- DOI: 10.1128/JVI.01501-08
Genetic history of hepatitis C virus in East Asia
Abstract
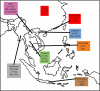
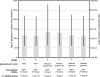
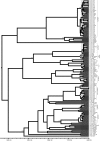
The hepatitis C virus (HCV), which currently infects an estimated 3% of people worldwide, has been present in some human populations for several centuries, notably HCV genotypes 1 and 2 in West Africa and genotype 6 in Southeast Asia. Here we use newly developed methods of sequence analysis to conduct the first comprehensive investigation of the epidemic and evolutionary history of HCV in Asia. Our analysis includes new HCV core (n = 16) and NS5B (n = 14) gene sequences, obtained from serum samples of jaundiced patients from Laos. These exceptionally diverse isolates were analyzed in conjunction with all available reference strains using phylogenetic and Bayesian coalescent methods. We performed statistical tests of phylogeographic structure and applied a recently developed "relaxed molecular clock" approach to HCV for the first time, which indicated an unexpectedly high degree of rate variation. Our results reveal a >1,000-year-long development of genotype 6 in Asia, characterized by substantial phylogeographic structure and two distinct phases of epidemic history, before and during the 20th century. We conclude that HCV lineages representing preexisting and spatially restricted strains were involved in multiple, independent local epidemics during the 20th century. Our analysis explains the generation and maintenance of HCV diversity in Asia and could provide a template for further investigations of HCV spread in other regions.
Figures



References
-
- Centers for Disease Control and Prevention. 1998. Recommendations for prevention and control of hepatitic C virus (HCV) infection and HCV-related chronic disease. MMWR Recomm. Rep. 47(RR-19)1-39. - PubMed
-
- Chen, Y. D., M. Y. Liu, W. L. Yu, J. Q. Li, M. Peng, Q. Dai, X. Liu, and Z. Q. Zhou. 2002. Hepatitis C virus infections and genotypes in China. Hepatobiliary Pancreat. Dis. Int. 1194-201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

