Identification of sequences in herpes simplex virus type 1 ICP22 that influence RNA polymerase II modification and viral late gene expression
- PMID: 18971282
- PMCID: PMC2612302
- DOI: 10.1128/JVI.01954-08
Identification of sequences in herpes simplex virus type 1 ICP22 that influence RNA polymerase II modification and viral late gene expression
Abstract
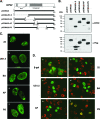
Previous studies have shown that the herpes simplex virus type 1 (HSV-1) immediate-early protein ICP22 alters the phosphorylation of the host cell RNA polymerase II (Pol II) during viral infection. In this study, we have engineered several ICP22 plasmid and virus mutants in order to map the ICP22 sequences that are involved in this function. We identify a region in the C-terminal half of ICP22 (residues 240 to 340) that is critical for Pol II modification and further show that the N-terminal half of the protein (residues 1 to 239) is not required. However, immunofluorescence analysis indicates that the N-terminal half of ICP22 is needed for its localization to nuclear body structures. These results demonstrate that ICP22's effects on Pol II do not require that it accumulate in nuclear bodies. As ICP22 is known to enhance viral late gene expression during infection of certain cultured cells, including human embryonic lung (HEL) cells, we used our engineered viral mutants to map this function of ICP22. It was found that mutations in both the N- and C-terminal halves of ICP22 result in similar defects in viral late gene expression and growth in HEL cells, despite having distinctly different effects on Pol II. Thus, our results genetically uncouple ICP22's effects on Pol II from its effects on viral late gene expression. This suggests that these two functions of ICP22 may be due to distinct activities of the protein.
Figures






References
-
- Advani, S. J., R. Brandimarti, R. R. Weichselbaum, and B. Roizman. 2000. The disappearance of cyclins A and B and the increase in activity of the G2/M-phase cellular kinase cdc2 in herpes simplex virus 1-infected cells require expression of the α22/US1.5 and UL13 viral genes. J. Virol. 748-15. - PMC - PubMed
-
- Cai, W. Z., S. Person, C. DebRoy, and B. H. Gu. 1988. Functional regions and structural features of the gB glycoprotein of herpes simplex virus type 1. An analysis of linker insertion mutants. J. Mol. Biol. 201575-588. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

