Evolution of MDA-5/RIG-I-dependent innate immunity: independent evolution by domain grafting
- PMID: 18971330
- PMCID: PMC2579374
- DOI: 10.1073/pnas.0804956105
Evolution of MDA-5/RIG-I-dependent innate immunity: independent evolution by domain grafting
Abstract
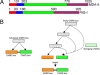
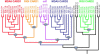
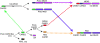
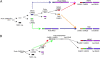
Type I Interferons (IFNs) are requisite components in antiviral innate immunity. Classically, a Toll-like receptor-dependent pathway induces type I interferons. However, recent recognition of melanoma differentiation associated gene-5 (MDA-5) and retinoic acid inducible gene-I (RIG-I) as primary sensors of RNA viruses for type I interferon induction highlights a potentially unique pathway for innate immunity. Our present investigation tracing the phylogenetic origin of MDA-5 and RIG-I domain arrangement (CARD1-CARD2-helicase-DEAD/DEAH) indicates that these proteins originated specifically in mammals, firmly linking this family of proteins with interferons in a highly derived evolutionary development of innate immunity. MDA-5, but not RIG-I, orthologs are found in fish, indicating that MDA-5 might have evolved before RIG-I. Our analyses also reveal that the MDA-5 and RIG-I domain arrangement evolved independently by domain grafting and not by a simple gene-duplication event of the entire four-domain arrangement, which may have been initiated by differential sensitivity of these proteins to viral infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Evolution of the DEAD box helicase family in chicken: chickens have no DHX9 ortholog.Microbiol Immunol. 2015 Oct;59(10):633-40. doi: 10.1111/1348-0421.12322. Microbiol Immunol. 2015. PMID: 26382053
-
[Non-self RNA-sensing mechanism of RIG-I RNA helicase].Seikagaku. 2008 Sep;80(9):838-43. Seikagaku. 2008. PMID: 18975620 Review. Japanese. No abstract available.
-
Origin and evolution of the RIG-I like RNA helicase gene family.BMC Evol Biol. 2009 Apr 28;9:85. doi: 10.1186/1471-2148-9-85. BMC Evol Biol. 2009. PMID: 19400936 Free PMC article.
-
Evolution of a Novel Antiviral Immune-Signaling Interaction by Partial-Gene Duplication.PLoS One. 2015 Sep 10;10(9):e0137276. doi: 10.1371/journal.pone.0137276. eCollection 2015. PLoS One. 2015. PMID: 26356745 Free PMC article.
-
[Structural and functional views of the intracellular viral RNA sensor RIG-I].Uirusu. 2008 Dec;58(2):97-103. doi: 10.2222/jsv.58.97. Uirusu. 2008. PMID: 19374188 Review. Japanese.
Cited by
-
Molecular cloning, characterization and expression analysis of woodchuck retinoic acid-inducible gene I.J Huazhong Univ Sci Technolog Med Sci. 2016 Jun;36(3):335-343. doi: 10.1007/s11596-016-1588-5. Epub 2016 Jul 5. J Huazhong Univ Sci Technolog Med Sci. 2016. PMID: 27376800
-
Comparative Structure and Function Analysis of the RIG-I-Like Receptors: RIG-I and MDA5.Front Immunol. 2019 Jul 17;10:1586. doi: 10.3389/fimmu.2019.01586. eCollection 2019. Front Immunol. 2019. PMID: 31379819 Free PMC article. Review.
-
A Glimpse of "Dicer Biology" Through the Structural and Functional Perspective.Front Mol Biosci. 2021 May 7;8:643657. doi: 10.3389/fmolb.2021.643657. eCollection 2021. Front Mol Biosci. 2021. PMID: 34026825 Free PMC article. Review.
-
Functional characterizations of RIG-I to GCRV and viral/bacterial PAMPs in grass carp Ctenopharyngodon idella.PLoS One. 2012;7(7):e42182. doi: 10.1371/journal.pone.0042182. Epub 2012 Jul 31. PLoS One. 2012. PMID: 22860079 Free PMC article.
-
Pattern recognition receptor MDA5 modulates CD8+ T cell-dependent clearance of West Nile virus from the central nervous system.J Virol. 2013 Nov;87(21):11401-15. doi: 10.1128/JVI.01403-13. Epub 2013 Aug 21. J Virol. 2013. PMID: 23966390 Free PMC article.
References
-
- Takeuchi O, Akira S. Recognition of viruses by innate immunity. Immunol Rev. 2007;220:214–224. - PubMed
-
- Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202(1):8–32. - PubMed
-
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

