Visualizing myosin-actin interaction with a genetically-encoded fluorescent strain sensor
- PMID: 18971336
- PMCID: PMC2579347
- DOI: 10.1073/pnas.0805513105
Visualizing myosin-actin interaction with a genetically-encoded fluorescent strain sensor
Abstract
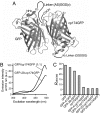
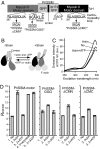
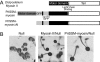
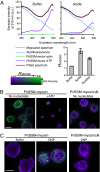
Many proteins have been shown to undergo conformational changes in response to externally applied force in vitro, but whether the force-induced protein conformational changes occur in vivo remains unclear. To reveal the force-induced conformational changes, or strains, within proteins in living cells, we have developed a genetically encoded fluorescent "strain sensor," by combining the proximity imaging (PRIM) technique, which uses spectral changes of 2 GFP molecules that are in direct contact, and myosin-actin as a model system. The developed PRIM-based strain sensor module (PriSSM) consists of the tandem fusion of a normal and circularly permuted GFP. To apply strain to PriSSM, it was inserted between 2 motor domains of Dictyostelium myosin II. In the absence of strain, the 2 GFP moieties in PriSSM are in contact, whereas when the motor domains are bound to F-actin, PriSSM has a strained conformation, leading to the loss of contact and a concomitant spectral change. Using the sensor system, we found that the position of the lever arm in the rigor state was affected by mutations within the motor domain. Moreover, the sensor was used to visualize the interaction between myosin II and F-actin in Dictyostelium cells. In normal cells, myosin was largely detached from F-actin, whereas ATP depletion or hyperosmotic stress increased the fraction of myosin bound to F-actin. The PRIM-based strain sensor may provide a general approach for studying force-induced protein conformational changes in cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Myosin-actin interaction in Dictyostelium cells revealed by GFP-based strain sensor and validated linear spectral unmixing.Cytometry A. 2010 Aug;77(8):743-50. doi: 10.1002/cyto.a.20900. Cytometry A. 2010. PMID: 20653014
-
Dictyostelium myosin II as a model to study the actin-myosin interactions during force generation.J Muscle Res Cell Motil. 2002;23(7-8):697-702. doi: 10.1023/a:1024415409406. J Muscle Res Cell Motil. 2002. PMID: 12952068 Review.
-
Stretching actin filaments within cells enhances their affinity for the myosin II motor domain.PLoS One. 2011;6(10):e26200. doi: 10.1371/journal.pone.0026200. Epub 2011 Oct 13. PLoS One. 2011. PMID: 22022566 Free PMC article.
-
Myosin II-dependent cylindrical protrusions induced by quinine in Dictyostelium: antagonizing effects of actin polymerization at the leading edge.J Cell Sci. 2001 Jun;114(Pt 11):2155-65. doi: 10.1242/jcs.114.11.2155. J Cell Sci. 2001. PMID: 11493651
-
Double-headed binding of myosin II to F-actin shows the effect of strain on head structure.J Struct Biol. 2023 Sep;215(3):107995. doi: 10.1016/j.jsb.2023.107995. Epub 2023 Jul 4. J Struct Biol. 2023. PMID: 37414375 Free PMC article.
Cited by
-
Orientation-based FRET sensor for real-time imaging of cellular forces.J Cell Sci. 2012 Feb 1;125(Pt 3):743-50. doi: 10.1242/jcs.093104. J Cell Sci. 2012. PMID: 22389408 Free PMC article.
-
Visualizing dynamic cytoplasmic forces with a compliance-matched FRET sensor.J Cell Sci. 2011 Jan 15;124(Pt 2):261-9. doi: 10.1242/jcs.071928. Epub 2010 Dec 20. J Cell Sci. 2011. PMID: 21172803 Free PMC article.
-
Molecular Force Sensors for Biological Application.Int J Mol Sci. 2024 Jun 4;25(11):6198. doi: 10.3390/ijms25116198. Int J Mol Sci. 2024. PMID: 38892386 Free PMC article. Review.
-
Enhanced Molecular Tension Sensor Based on Bioluminescence Resonance Energy Transfer (BRET).ACS Sens. 2020 Jan 24;5(1):34-39. doi: 10.1021/acssensors.9b00796. Epub 2020 Jan 8. ACS Sens. 2020. PMID: 31872754 Free PMC article.
-
Opticool: Cutting-edge transgenic optical tools.PLoS Genet. 2024 Mar 22;20(3):e1011208. doi: 10.1371/journal.pgen.1011208. eCollection 2024 Mar. PLoS Genet. 2024. PMID: 38517915 Free PMC article. Review.
References
-
- Forman JR, Clarke J. Mechanical unfolding of proteins: Insights into biology, structure, and folding. Curr Opin Struct Biol. 2007;17:58–66. - PubMed
-
- Tskhovrebova L, Trinick J, Sleep JA, Simmons RM. Elasticity and unfolding of single molecules of the giant muscle protein titin. Nature. 1997;387:308–312. - PubMed
-
- Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. - PubMed
-
- Kellermayer MS, Smith SB, Granzier HL, Bustamante C. Folding–unfolding transitions in single titin molecules characterized with laser tweezers. Science. 1997;276:1112–1116. - PubMed
-
- Rief M, Pascual J, Saraste M, Gaub HE. Single-molecule force spectroscopy of spectrin repeats: Low unfolding forces in helix bundles. J Mol Biol. 1999;286:553–561. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

