Control of mRNA decapping by Dcp2: An open and shut case?
- PMID: 18971632
- PMCID: PMC2699264
- DOI: 10.4161/rna.6859
Control of mRNA decapping by Dcp2: An open and shut case?
Abstract
mRNA decapping by Dcp2 is a critical step in several major eukaryotic mRNA decay pathways. Dcp2 forms the catalytic core of a mRNP that is configured for processing diverse substrates by pathway-specific activators. Here we elaborate a model of catalysis by Dcp2 which posits that activity is controlled by a conformational equilibrium between an open, inactive and closed, active form of the enzyme. Structural studies on yeast Dcp2 indicate that the general activator Dcp1 and substrate promote the closed form of the enzyme. Kinetic studies indicate the catalytic step of decapping is rate-limiting and accelerated by Dcp1. We propose that regulation of conformational transitions in Dcp2 during a rate-limiting step after assembly of the decapping mRNP provides a checkpoint for determining if an mRNA is degraded or recycled to translation.
Figures

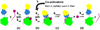
References
-
- Schier AF. The maternal-zygotic transition: death and birth of RNAs. Science. 2007;316:406–407. - PubMed
-
- Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. - PubMed
-
- Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5:89–99. - PubMed
-
- Amrani N, Sachs MS, Jacobson A. Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol. 2006;7:415–425. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
