Low reduction potential of Ero1alpha regulatory disulphides ensures tight control of substrate oxidation
- PMID: 18971943
- PMCID: PMC2579255
- DOI: 10.1038/emboj.2008.230
Low reduction potential of Ero1alpha regulatory disulphides ensures tight control of substrate oxidation
Abstract
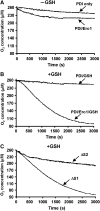
Formation of disulphide bonds within the mammalian endoplasmic reticulum (ER) requires the combined activities of Ero1alpha and protein disulphide isomerase (PDI). As Ero1alpha produces hydrogen peroxide during oxidation, regulation of its activity is critical in preventing ER-generated oxidative stress. Here, we have expressed and purified recombinant human Ero1alpha and shown that it has activity towards thioredoxin and PDI. The activity towards PDI required the inclusion of glutathione to ensure sustained oxidation. By carrying out site-directed mutagenesis of cysteine residues, we show that Ero1alpha is regulated by non-catalytic disulphides. The midpoint reduction potential (E degrees') of the regulatory disulphides was calculated to be approximately -275 mV making them stable in the redox conditions prevalent in the ER. The stable regulatory disulphides were only partially reduced by PDI (E degrees' approximately -180 mV), suggesting either that this is a mechanism for preventing excessive Ero1alpha activity and oxidation of PDI or that additional factors are required for Ero1alpha activation within the mammalian ER.
Figures

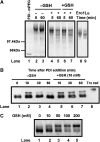

 ) or regulatory disulphides (
) or regulatory disulphides ( ). Known disulphide pairings are as indicated as they exist in the inactive state of the protein. Note that in the human protein cys94 and cys99 are engaged in disulphide pairings with cys131 and cys104, respectively, whereas in the active state these would form a disulphide with each other.
). Known disulphide pairings are as indicated as they exist in the inactive state of the protein. Note that in the human protein cys94 and cys99 are engaged in disulphide pairings with cys131 and cys104, respectively, whereas in the active state these would form a disulphide with each other.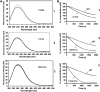

Comment in
-
A non-catalytic disulphide bond regulating redox flux in the ER oxidative folding pathway.EMBO J. 2009 Feb 4;28(3):169-70. doi: 10.1038/emboj.2008.293. EMBO J. 2009. PMID: 19194483 Free PMC article. No abstract available.
References
-
- Appenzeller-Herzog C, Ellgaard L (2008) In vivo reduction–oxidation state of protein disulphide isomerase: the two active sites independently occur in the reduced and oxidized forms. Antioxid Redox Signal 10: 55–64 - PubMed
-
- Bertoli G, Simmen T, Anelli T, Molteni SN, Fesce R, Sitia R (2004) Two conserved cysteine triads in human Ero1alpha cooperate for efficient disulphide bond formation in the endoplasmic reticulum. J Biol Chem 279: 30047–30052 - PubMed
-
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72: 248–254 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

