Factors affecting human supragingival biofilm composition. I. Plaque mass
- PMID: 18973540
- PMCID: PMC2710401
- DOI: 10.1111/j.1600-0765.2008.01154.x
Factors affecting human supragingival biofilm composition. I. Plaque mass
Abstract
Background and objective: Little is known about the factors that affect the microbial composition of supragingival biofilms. This study was designed to examine the relationship between total DNA probe counts of supragingival biofilm samples, clinical parameters and supragingival biofilm composition.
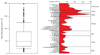
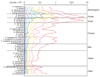
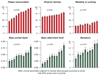
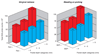
Material and methods: Supragingival plaque samples were taken from 187 systemically healthy adult subjects (n = 4745 samples). All samples were individually analyzed for their content of 40 bacterial species using checkerboard DNA-DNA hybridization. The relationship between total DNA probe counts and microbial composition was examined by subsetting the data into 10 groups based on 10 percentile increments of the total DNA probe counts. Differences among groups in terms of species counts and proportions were sought, as well as relationships of total plaque DNA probe count and clinical parameters.
Results: There was a wide distribution in mean total DNA probe counts among the 187 subjects. With increasing total plaque levels there was a change in the proportions of individual species and microbial complexes. 'Small plaques' were characterized by high proportions of species in the yellow, orange, purple and 'other' complexes; plaques of moderate mass were characterized by high proportions of Actinomyces and purple complex species, while 'large plaques' exhibited increased proportions of green and orange complex species. Measures of gingival inflammation, pocket depth and recession were significantly positively associated with total DNA probe counts. Increased plaque numbers were related to increased pocket depth irrespective of presence or absence of gingival inflammation.
Conclusion: The proportions of individual species and microbial complexes in supragingival biofilms are influenced by the total numbers of organisms in the biofilm.
Figures




References
-
- Sharawy AM, Sabharwal K, Socransky SS, Lobene RR. A quantitative study of plaque and calculus formation in normal and periodontally involved mouths. J Periodontol. 1966;37:495–501. - PubMed
-
- Zee KY, Samaranayake LP, Attstrom R. Predominant cultivable supragingival plaque in Chinese “rapid” and “slow” plaque formers. J Clin Periodontol. 1996;23:1025–1031. - PubMed
-
- Socransky SS, Smith C, Martin L, Paster BJ, Dewhirst FE, Levin AE. “Checkerboard” DNA-DNA hybridization. Biotechniques. 1994;17:788–792. - PubMed
-
- Socransky SS, Haffajee AD, Smith C, Martin L, Haffajee JA, Uzel NG, et al. The use of checkerboard DNA-DNA hybridization to study complex microbial ecosystems. Oral Microbiol Immunol. 2004;19:352–362. - PubMed
-
- Ludwig JA, Reynolds JF. A primer on methods and computing. New York: Wiley; 1988. Statistical ecology; pp. 89–95.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

