Towards the understanding of the cocoa transcriptome: Production and analysis of an exhaustive dataset of ESTs of Theobroma cacao L. generated from various tissues and under various conditions
- PMID: 18973681
- PMCID: PMC2642826
- DOI: 10.1186/1471-2164-9-512
Towards the understanding of the cocoa transcriptome: Production and analysis of an exhaustive dataset of ESTs of Theobroma cacao L. generated from various tissues and under various conditions
Abstract
Background: Theobroma cacao L., is a tree originated from the tropical rainforest of South America. It is one of the major cash crops for many tropical countries. T. cacao is mainly produced on smallholdings, providing resources for 14 million farmers. Disease resistance and T. cacao quality improvement are two important challenges for all actors of cocoa and chocolate production. T. cacao is seriously affected by pests and fungal diseases, responsible for more than 40% yield losses and quality improvement, nutritional and organoleptic, is also important for consumers. An international collaboration was formed to develop an EST genomic resource database for cacao.
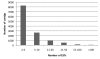
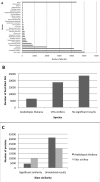
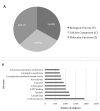
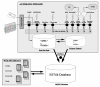
Results: Fifty-six cDNA libraries were constructed from different organs, different genotypes and different environmental conditions. A total of 149,650 valid EST sequences were generated corresponding to 48,594 unigenes, 12,692 contigs and 35,902 singletons. A total of 29,849 unigenes shared significant homology with public sequences from other species.Gene Ontology (GO) annotation was applied to distribute the ESTs among the main GO categories.A specific information system (ESTtik) was constructed to process, store and manage this EST collection allowing the user to query a database.To check the representativeness of our EST collection, we looked for the genes known to be involved in two different metabolic pathways extensively studied in other plant species and important for T. cacao qualities: the flavonoid and the terpene pathways. Most of the enzymes described in other crops for these two metabolic pathways were found in our EST collection.A large collection of new genetic markers was provided by this ESTs collection.
Conclusion: This EST collection displays a good representation of the T. cacao transcriptome, suitable for analysis of biochemical pathways based on oligonucleotide microarrays derived from these ESTs. It will provide numerous genetic markers that will allow the construction of a high density gene map of T. cacao. This EST collection represents a unique and important molecular resource for T. cacao study and improvement, facilitating the discovery of candidate genes for important T. cacao trait variation.
Figures



References
-
- Figueira A, Janik J, Goldsbrough P. Genome size and DNA polymorphism in Theobroma cacao. Journal of the American Society for Horticultural Science. 1992;117:673–677.
-
- Lanaud C, Hamon P, Duperray C. Estimation of nuclear DNA content of Theobroma cacao L. by flow cytometry. Café, Cacao, Thé. 1992;36:3–8.
-
- Cheesman EE. Notes on the nomenclature, classification possible and relationships of cocoa populations. Tropical Agriculture. 1944;21:144–159.
-
- Bowers JH, Bailey BA, Hebbar PK, Sanogo S, Lumsden RD. The impact of plant diseases on world chocolate production. Plant Health Progress. 2001.
-
- Ampuero E. Monilia pod rot of cocoa. Cocoa Grower's Bulletin. 1967;9:1518.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

