Genomic plasticity of the immune-related Mhc class I B region in macaque species
- PMID: 18973694
- PMCID: PMC2584111
- DOI: 10.1186/1471-2164-9-514
Genomic plasticity of the immune-related Mhc class I B region in macaque species
Abstract
Background: In sharp contrast to humans and great apes, the expanded Mhc-B region of rhesus and cynomolgus macaques is characterized by the presence of differential numbers and unique combinations of polymorphic class I B genes per haplotype. The MIB microsatellite is closely linked to the single class I B gene in human and in some great apes studied. The physical map of the Mhc of a heterozygous rhesus monkey provides unique material to analyze MIB and Mamu-B copy number variation and then allows one to decipher the compound evolutionary history of this region in primate species.
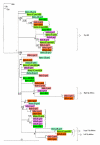
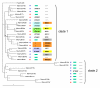
Results: In silico research pinpointed 12 MIB copies (duplicons), most of which are associated with expressed B-genes that cluster in a separate clade in the phylogenetic tree. Generic primers tested on homozygous rhesus and pedigreed cynomolgus macaques allowed the identification of eight to eleven MIB copies per individual. The number of MIB copies present per haplotype varies from a minimum of three to six in cynomolgus macaques and from five to eight copies in rhesus macaques. Phylogenetic analyses highlight a strong transpecific sharing of MIB duplicons. Using the physical map, we observed that, similar to MIB duplicons, highly divergent Mamu-B genes can be present on the same haplotype. Haplotype variation as reflected by the copy number variation of class I B loci is best explained by recombination events, which are found to occur between MIBs and Mamu-B.
Conclusion: The data suggest the existence of highly divergent MIB and Mamu-B lineages on a given haplotype, as well as variable MIB and B copy numbers and configurations, at least in rhesus macaque. Recombination seems to occur between MIB and Mamu-B loci, and the resulting haplotypic plasticity at the individual level may be a strategy to better cope with pathogens. Therefore, evolutionary inferences based on the multiplicated MIB loci but also other markers close to B-genes appear to be promising for the study of B-region organization and evolution in primates.
Figures

References
-
- Lagaaij EL, Hennemann IP, Ruigrok M, de Haan MW, Persijn GG, Termijtelen A, Hendricks GF, Weimar W, Claas FH, van Rood JJ. Effect of one-HLA-DR-antigen-matched and completely HLA-DR-mismatched blood transfusions on survival of heart and kidney allografts. N Engl J Med. 1989;321:701–705. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

