EWS-FLI1 induces developmental abnormalities and accelerates sarcoma formation in a transgenic mouse model
- PMID: 18974141
- PMCID: PMC4167779
- DOI: 10.1158/0008-5472.CAN-08-0573
EWS-FLI1 induces developmental abnormalities and accelerates sarcoma formation in a transgenic mouse model
Abstract
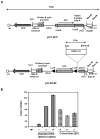
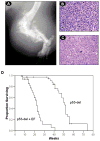
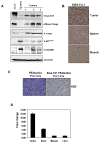
Ewing's sarcoma is characterized by the t(11;22)(q24:q12) reciprocal translocation. To study the effects of the fusion gene EWS-FLI1 on development and tumor formation, a transgenic mouse model was created. A strategy of conditional expression was used to limit the potentially deleterious effects of EWS-FLI1 to certain tissues. In the absence of Cre recombinase, EWS-FLI1 was not expressed in the EWS-FLI1 transgenic mice, and they had a normal phenotype. When crossed to the Prx1-Cre transgenic mouse, which expresses Cre recombinase in the primitive mesenchymal cells of the embryonic limb bud, the EF mice were noted to have a number of developmental defects of the limbs. These included shortening of the limbs, muscle atrophy, cartilage dysplasia, and immature bone. By itself, EWS-FLI1 did not induce the formation of tumors in the EF transgenic mice. However, in the setting of p53 deletion, EWS-FLI1 accelerated the formation of sarcomas from a median time of 50 to 21 weeks. Furthermore, EWS-FLI1 altered the type of tumor that formed. Conditional deletion of p53 in mesenchymal cells (Prx1-Cre p53(lox/lox)) produced osteosarcomas as the predominant tumor. The presence of EWS-FLI1 shifted the tumor phenotype to a poorly differentiated sarcoma. The results taken together suggest that EWS-FLI1 inhibits normal limb development and accelerates the formation of poorly differentiated sarcomas.
Figures


Similar articles
-
Differential transactivation by alternative EWS-FLI1 fusion proteins correlates with clinical heterogeneity in Ewing's sarcoma.Cancer Res. 1999 Apr 1;59(7):1428-32. Cancer Res. 1999. PMID: 10197607
-
EWS-FLI1 suppresses NOTCH-activated p53 in Ewing's sarcoma.Cancer Res. 2008 Sep 1;68(17):7100-9. doi: 10.1158/0008-5472.CAN-07-6145. Cancer Res. 2008. PMID: 18757425 Free PMC article.
-
Identification of various exon combinations of the ews/fli1 translocation: an optimized RT-PCR method for paraffin embedded tissue -- a report by the CWS-study group.Klin Padiatr. 2004 Nov-Dec;216(6):315-22. doi: 10.1055/s-2004-832338. Klin Padiatr. 2004. PMID: 15565546
-
EWS-FLI1 and Ewing's sarcoma: recent molecular data and new insights.Cancer Biol Ther. 2002 Jul-Aug;1(4):330-6. Cancer Biol Ther. 2002. PMID: 12432241 Review.
-
Challenges in modeling EWS-FLI1-driven transgenic mouse model for Ewing sarcoma.Am J Transl Res. 2021 Nov 15;13(11):12181-12194. eCollection 2021. Am J Transl Res. 2021. PMID: 34956445 Free PMC article. Review.
Cited by
-
Loss of Stag2 cooperates with EWS-FLI1 to transform murine Mesenchymal stem cells.BMC Cancer. 2020 Jan 2;20(1):3. doi: 10.1186/s12885-019-6465-8. BMC Cancer. 2020. PMID: 31898537 Free PMC article.
-
Efficacy of ATR inhibitors as single agents in Ewing sarcoma.Oncotarget. 2016 Sep 13;7(37):58759-58767. doi: 10.18632/oncotarget.11643. Oncotarget. 2016. PMID: 27577084 Free PMC article.
-
Dr. Jekyll and Mr. Hyde: The Two Faces of the FUS/EWS/TAF15 Protein Family.Sarcoma. 2011;2011:837474. doi: 10.1155/2011/837474. Epub 2010 Dec 9. Sarcoma. 2011. PMID: 21197473 Free PMC article.
-
Preclinical models for the study of pediatric solid tumors: focus on bone sarcomas.Front Oncol. 2024 Jul 18;14:1388484. doi: 10.3389/fonc.2024.1388484. eCollection 2024. Front Oncol. 2024. PMID: 39091911 Free PMC article. Review.
-
EWS/FLI utilizes NKX2-2 to repress mesenchymal features of Ewing sarcoma.Genes Cancer. 2015 Mar;6(3-4):129-43. doi: 10.18632/genesandcancer.57. Genes Cancer. 2015. PMID: 26000096 Free PMC article.
References
-
- Delattre O, Zucman J, Plougastel B, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–5. - PubMed
-
- Dejana E. Endothelial cell-cell junctions: happy together. Nat Rev Mol Cell Biol. 2004;5:261–70. - PubMed
-
- Rettig WJ, Garin-Chesa P, Huvos AG. Ewing’s sarcoma: new approaches to histogenesis and molecular plasticity. Lab Invest. 1992;66:133–7. - PubMed
-
- Ushigome S, Machinami R, Sorensen PH. Ewingsarcoma/primitive neuroectodermal tumor. In: Fletcher CDM, Unni KK, Mertens F, editors. WHO Classification ofTumors, Pathology and Genetics, Tumors of Soft Tissue and Bone. 2003.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

