The secondary structure of the 5' end of the FIV genome reveals a long-range interaction between R/U5 and gag sequences, and a large, stable stem-loop
- PMID: 18974279
- PMCID: PMC2590967
- DOI: 10.1261/rna.1284908
The secondary structure of the 5' end of the FIV genome reveals a long-range interaction between R/U5 and gag sequences, and a large, stable stem-loop
Abstract
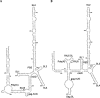
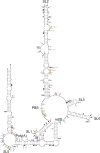
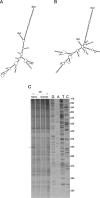
Feline immunodeficiency virus (FIV) is a lentivirus that infects cats and is related to human immunodeficiency virus (HIV). Although it is a common worldwide infection, and has potential uses as a human gene therapy vector and as a nonprimate model for HIV infection, little detail is known of the viral life cycle. Previous experiments have shown that its packaging signal includes two or more regions within the first 511 nucleotides of the genomic RNA. We have undertaken a secondary structural analysis of this RNA by minimal free-energy structural prediction, biochemical mapping, and phylogenetic analysis, and show that it contains five conserved stem-loops and a conserved long-range interaction between heptanucleotide sequences 5'-CCCUGUC-3' in R/U5 and 5'-GACAGGG-3' in gag. This long-range interaction is similar to that seen in primate lentiviruses where it is thought to be functionally important. Along with strains that infect domestic cats, this heptanucleotide interaction can also occur in species-specific FIV strains that infect pumas, lions, and Pallas' cats where the heptanucleotide sequences involved vary. We have analyzed spliced and genomic FIV RNAs and see little structural change or sequence conservation within single-stranded regions of the 5' UTR that are important for viral packaging, suggesting that FIV may employ a cotranslational packaging mechanism.
Figures



Similar articles
-
Optimal packaging of FIV genomic RNA depends upon a conserved long-range interaction and a palindromic sequence within gag.J Mol Biol. 2010 Oct 15;403(1):103-119. doi: 10.1016/j.jmb.2010.08.019. Epub 2010 Aug 21. J Mol Biol. 2010. PMID: 20732330 Free PMC article.
-
Mapping the encapsidation determinants of feline immunodeficiency virus.J Virol. 2002 Dec;76(23):11889-903. doi: 10.1128/jvi.76.23.11889-11903.2002. J Virol. 2002. PMID: 12414931 Free PMC article.
-
SHAPE analysis of the FIV Leader RNA reveals a structural switch potentially controlling viral packaging and genome dimerization.Nucleic Acids Res. 2011 Aug;39(15):6692-704. doi: 10.1093/nar/gkr252. Epub 2011 May 5. Nucleic Acids Res. 2011. PMID: 21546549 Free PMC article.
-
The genome of feline immunodeficiency virus.Arch Virol. 1994;134(3-4):221-34. doi: 10.1007/BF01310563. Arch Virol. 1994. PMID: 8129613 Review.
-
Molecular properties of feline immunodeficiency virus (FIV).Infect Agents Dis. 1993 Dec;2(6):361-74. Infect Agents Dis. 1993. PMID: 8012737 Review. No abstract available.
Cited by
-
Stabilizing role of structural elements within the 5´ Untranslated Region (UTR) and gag sequences in Mason-Pfizer monkey virus (MPMV) genomic RNA packaging.RNA Biol. 2019 May;16(5):612-625. doi: 10.1080/15476286.2019.1572424. Epub 2019 Feb 17. RNA Biol. 2019. PMID: 30773097 Free PMC article.
-
New windows into retroviral RNA structures.Retrovirology. 2018 Jan 25;15(1):11. doi: 10.1186/s12977-018-0393-6. Retrovirology. 2018. PMID: 29368653 Free PMC article. Review.
-
Probing Retroviral and Retrotransposon Genome Structures: The "SHAPE" of Things to Come.Mol Biol Int. 2012;2012:530754. doi: 10.1155/2012/530754. Epub 2012 May 17. Mol Biol Int. 2012. PMID: 22685659 Free PMC article.
-
Properties and Functions of Feline Immunodeficiency Virus Gag Domains in Virion Assembly and Budding.Viruses. 2018 May 16;10(5):261. doi: 10.3390/v10050261. Viruses. 2018. PMID: 29772651 Free PMC article. Review.
-
Structural basis of genomic RNA (gRNA) dimerization and packaging determinants of mouse mammary tumor virus (MMTV).Retrovirology. 2014 Nov 14;11:96. doi: 10.1186/s12977-014-0096-6. Retrovirology. 2014. PMID: 25394412 Free PMC article.
References
-
- Abbink T.E., Berkhout B. A novel long-distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J. Biol. Chem. 2003;278:11601–11611. - PubMed
-
- Browning M.T., Mustafa F., Schmidt R.D., Lew K.A., Rizvi T.A. Delineation of sequences important for efficient packaging of feline immunodeficiency virus RNA. J. Gen. Virol. 2003a;84:621–627. - PubMed
-
- Browning M.T., Mustafa F., Schmidt R.D., Lew K.A., Rizvi T.A. Sequences within the gag gene of feline immunodeficiency virus (FIV) are important for efficient RNA encapsidation. Virus Res. 2003b;93:199–209. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
