Structural evidence for common ancestry of the nuclear pore complex and vesicle coats
- PMID: 18974315
- PMCID: PMC2680690
- DOI: 10.1126/science.1165886
Structural evidence for common ancestry of the nuclear pore complex and vesicle coats
Abstract
Nuclear pore complexes (NPCs) facilitate nucleocytoplasmic transport. These massive assemblies comprise an eightfold symmetric scaffold of architectural proteins and central-channel phenylalanine-glycine-repeat proteins forming the transport barrier. We determined the nucleoporin 85 (Nup85)*Seh1 structure, a module in the heptameric Nup84 complex, at 3.5 angstroms resolution. Structural, biochemical, and genetic analyses position the Nup84 complex in two peripheral NPC rings. We establish a conserved tripartite element, the ancestral coatomer element ACE1, that reoccurs in several nucleoporins and vesicle coat proteins, providing structural evidence of coevolution from a common ancestor. We identified interactions that define the organization of the Nup84 complex on the basis of comparison with vesicle coats and confirmed the sites by mutagenesis. We propose that the NPC scaffold, like vesicle coats, is composed of polygons with vertices and edges forming a membrane-proximal lattice that provides docking sites for additional nucleoporins.
Figures



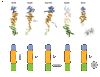
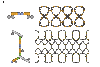
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

