Halothiobacillus neapolitanus carboxysomes sequester heterologous and chimeric RubisCO species
- PMID: 18974784
- PMCID: PMC2570492
- DOI: 10.1371/journal.pone.0003570
Halothiobacillus neapolitanus carboxysomes sequester heterologous and chimeric RubisCO species
Abstract
Background: The carboxysome is a bacterial microcompartment that consists of a polyhedral protein shell filled with ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO), the enzyme that catalyzes the first step of CO2 fixation via the Calvin-Benson-Bassham cycle.
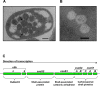
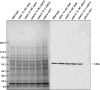
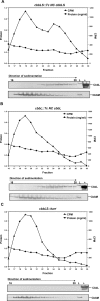
Methodology/principal findings: To analyze the role of RubisCO in carboxysome biogenesis in vivo we have created a series of Halothiobacillus neapolitanus RubisCO mutants. We identified the large subunit of the enzyme as an important determinant for its sequestration into alpha-carboxysomes and found that the carboxysomes of H. neapolitanus readily incorporate chimeric and heterologous RubisCO species. Intriguingly, a mutant lacking carboxysomal RubisCO assembles empty carboxysome shells of apparently normal shape and composition.
Conclusions/significance: These results indicate that carboxysome shell architecture is not determined by the enzyme they normally sequester. Our study provides, for the first time, clear evidence that carboxysome contents can be manipulated and suggests future nanotechnological applications that are based upon engineered protein microcompartments.
Conflict of interest statement
Figures




References
-
- Shively JM, editor. Berlin/Heidelberg: Springer; 2006. Complex Intracellular Structures in Prokaryotes.
-
- Heinhorst S, Cannon GC, Shively JM. Carboxysomes and carboxysome-like inclusions. In: Shively JM, editor. Complex Intracellular Structures in Prokaryotes. Berlin/Heidelberg: Springer; 2006. pp. 141–164.
-
- Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, et al. Protein structures forming the shell of primitive bacterial organelles. Science. 2005;309:936–938. - PubMed
-
- Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, et al. Atomic-level models of the bacterial carboxysome shell. Science. 2008;319:1083–1086. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

