Silencing and nuclear repositioning of the lambda5 gene locus at the pre-B cell stage requires Aiolos and OBF-1
- PMID: 18974788
- PMCID: PMC2571989
- DOI: 10.1371/journal.pone.0003568
Silencing and nuclear repositioning of the lambda5 gene locus at the pre-B cell stage requires Aiolos and OBF-1
Abstract
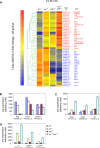
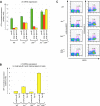
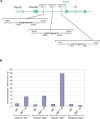
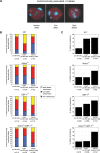
The chromatin regulator Aiolos and the transcriptional coactivator OBF-1 have been implicated in regulating aspects of B cell maturation and activation. Mice lacking either of these factors have a largely normal early B cell development. However, when both factors are eliminated simultaneously a block is uncovered at the transition between pre-B and immature B cells, indicating that these proteins exert a critical function in developing B lymphocytes. In mice deficient for Aiolos and OBF-1, the numbers of immature B cells are reduced, small pre-BII cells are increased and a significant impairment in immunoglobulin light chain DNA rearrangement is observed. We identified genes whose expression is deregulated in the pre-B cell compartment of these mice. In particular, we found that components of the pre-BCR, such as the surrogate light chain genes lambda5 and VpreB, fail to be efficiently silenced in double-mutant mice. Strikingly, developmentally regulated nuclear repositioning of the lambda5 gene is impaired in pre-B cells lacking OBF-1 and Aiolos. These studies uncover a novel role for OBF-1 and Aiolos in controlling the transcription and nuclear organization of genes involved in pre-BCR function.
Conflict of interest statement
Figures



Similar articles
-
Lack of the transcriptional coactivator OBF-1 prevents the development of systemic lupus erythematosus-like phenotypes in Aiolos mutant mice.J Immunol. 2003 Feb 15;170(4):1699-706. doi: 10.4049/jimmunol.170.4.1699. J Immunol. 2003. PMID: 12574333
-
Lambda5 is required for rearrangement of the Ig kappa light chain gene in pro-B cell lines.Int Immunol. 1999 Aug;11(8):1195-202. doi: 10.1093/intimm/11.8.1195. Int Immunol. 1999. PMID: 10421777
-
CD22 regulates early B cell development in BOB.1/OBF.1-deficient mice.Eur J Immunol. 2002 Sep;32(9):2481-9. doi: 10.1002/1521-4141(200209)32:9<2481::AID-IMMU2481>3.0.CO;2-C. Eur J Immunol. 2002. PMID: 12207332
-
Fidelity and infidelity in commitment to B-lymphocyte lineage development.Immunol Rev. 2000 Jun;175:104-11. Immunol Rev. 2000. PMID: 10933595 Review.
-
The pre-B cell receptor and its role in proliferation and Ig heavy chain allelic exclusion.Semin Immunol. 2002 Oct;14(5):335-42. doi: 10.1016/s1044-5323(02)00066-0. Semin Immunol. 2002. PMID: 12220934 Review.
Cited by
-
AIOLOS-Associated Inborn Errors of Immunity.J Clin Immunol. 2024 May 22;44(6):128. doi: 10.1007/s10875-024-01730-9. J Clin Immunol. 2024. PMID: 38773004 Free PMC article. Review.
-
Interchromosomal association and gene regulation in trans.Trends Genet. 2010 Apr;26(4):188-97. doi: 10.1016/j.tig.2010.01.007. Epub 2010 Mar 16. Trends Genet. 2010. PMID: 20236724 Free PMC article. Review.
-
Chronic lymphocytic leukaemia: could immunological tolerance mechanisms be the origin of lymphoid neoplasms?Immunology. 2014 Aug;142(4):536-50. doi: 10.1111/imm.12285. Immunology. 2014. PMID: 24645778 Free PMC article. Review.
-
Combined immunofluorescence and DNA FISH on 3D-preserved interphase nuclei to study changes in 3D nuclear organization.J Vis Exp. 2013 Feb 3;(72):e50087. doi: 10.3791/50087. J Vis Exp. 2013. PMID: 23407477 Free PMC article.
-
Stage-specific control of early B cell development by the transcription factor Ikaros.Nat Immunol. 2014 Mar;15(3):283-93. doi: 10.1038/ni.2828. Epub 2014 Feb 9. Nat Immunol. 2014. PMID: 24509509 Free PMC article.
References
-
- Busslinger M. Transcriptional control of early B cell development. Annu Rev Immunol. 2004;22:55–79. - PubMed
-
- Johnson K, Shapiro-Shelef M, Tunyaplin C, Calame K. Regulatory events in early and late B-cell differentiation. Molecular Immunology. 2005;42:749. - PubMed
-
- Matthias P, Rolink AG. Transcriptional networks in developing and mature B cells. Nature Reviews Immunology. 2005;5:497. - PubMed
-
- Hendriks RW, Middendorp S. The pre-BCR checkpoint as a cell-autonomous proliferation switch. Trends in Immunology. 2004;25:249. - PubMed
-
- ten Boekel E, Yamagami T, Andersson J, Rolink AG, Melchers F. The formation and selection of cells expressing preB cell receptors and B cell receptors. Curr Top Microbiol Immunol. 1999;246:3–9. discussion 9–10. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

