Direct identification of the Meloidogyne incognita secretome reveals proteins with host cell reprogramming potential
- PMID: 18974830
- PMCID: PMC2568823
- DOI: 10.1371/journal.ppat.1000192
Direct identification of the Meloidogyne incognita secretome reveals proteins with host cell reprogramming potential
Abstract
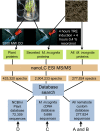
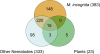
The root knot nematode, Meloidogyne incognita, is an obligate parasite that causes significant damage to a broad range of host plants. Infection is associated with secretion of proteins surrounded by proliferating cells. Many parasites are known to secrete effectors that interfere with plant innate immunity, enabling infection to occur; they can also release pathogen-associated molecular patterns (PAMPs, e.g., flagellin) that trigger basal immunity through the nematode stylet into the plant cell. This leads to suppression of innate immunity and reprogramming of plant cells to form a feeding structure containing multinucleate giant cells. Effectors have generally been discovered using genetics or bioinformatics, but M. incognita is non-sexual and its genome sequence has not yet been reported. To partially overcome these limitations, we have used mass spectrometry to directly identify 486 proteins secreted by M. incognita. These proteins contain at least segmental sequence identity to those found in our 3 reference databases (published nematode proteins; unpublished M. incognita ESTs; published plant proteins). Several secreted proteins are homologous to plant proteins, which they may mimic, and they contain domains that suggest known effector functions (e.g., regulating the plant cell cycle or growth). Others have regulatory domains that could reprogram cells. Using in situ hybridization we observed that most secreted proteins were produced by the subventral glands, but we found that phasmids also secreted proteins. We annotated the functions of the secreted proteins and classified them according to roles they may play in the development of root knot disease. Our results show that parasite secretomes can be partially characterized without cognate genomic DNA sequence. We observed that the M. incognita secretome overlaps the reported secretome of mammalian parasitic nematodes (e.g., Brugia malayi), suggesting a common parasitic behavior and a possible conservation of function between metazoan parasites of plants and animals.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Sasser JN, Eisenback JD, Carter CC, Triantaphyllou AC. THE INTERNATIONAL MELOIDOGYNE PROJECT - ITS GOALS AND ACCOMPLISHMENTS. Annual Review of Phytopathology. 1983;21:271–288.
-
- Caillaud MC, Dubreuil G, Quentin M, Perfus-Barbeoch L, Lecornte P, et al. Root-knot nematodes manipulate plant cell functions during a compatible interaction. Journal of Plant Physiology. 2008;165(1):104–113. - PubMed
-
- Panstruga R. Establishing compatibility between plants and obligate biotrophic pathogens. Current Opinion in Plant Biology. 2003;6(4):320–326. - PubMed
-
- Hussey RS. Disease-Inducing Secretions of Plant-Parasitic Nematodes. Annual Review of Phytopathology. 1989;27(1):123–141.
-
- Wyss U, Zunke U. Observations on the behaviour of second stage juveniles of Heterodera schachtii inside host roots. Revue de Nematologie. 1986;9(2):153–165.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

