A novel gene signature for molecular diagnosis of human prostate cancer by RT-qPCR
- PMID: 18974881
- PMCID: PMC2570792
- DOI: 10.1371/journal.pone.0003617
A novel gene signature for molecular diagnosis of human prostate cancer by RT-qPCR
Abstract
Background: Prostate cancer (CaP) is one of the most relevant causes of cancer death in Western Countries. Although detection of CaP at early curable stage is highly desirable, actual screening methods present limitations and new molecular approaches are needed. Gene expression analysis increases our knowledge about the biology of CaP and may render novel molecular tools, but the identification of accurate biomarkers for reliable molecular diagnosis is a real challenge. We describe here the diagnostic power of a novel 8-genes signature: ornithine decarboxylase (ODC), ornithine decarboxylase antizyme (OAZ), adenosylmethionine decarboxylase (AdoMetDC), spermidine/spermine N(1)-acetyltransferase (SSAT), histone H3 (H3), growth arrest specific gene (GAS1), glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and Clusterin (CLU) in tumour detection/classification of human CaP.
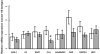
Methodology/principal findings: The 8-gene signature was detected by retrotranscription real-time quantitative PCR (RT-qPCR) in frozen prostate surgical specimens obtained from 41 patients diagnosed with CaP and recommended to undergo radical prostatectomy (RP). No therapy was given to patients at any time before RP. The bio-bank used for the study consisted of 66 specimens: 44 were benign-CaP paired from the same patient. Thirty-five were classified as benign and 31 as CaP after final pathological examination. Only molecular data were used for classification of specimens. The Nearest Neighbour (NN) classifier was used in order to discriminate CaP from benign tissue. Validation of final results was obtained with 10-fold cross-validation procedure. CaP versus benign specimens were discriminated with (80+/-5)% accuracy, (81+/-6)% sensitivity and (78+/-7)% specificity. The method also correctly classified 71% of patients with Gleason score<7 versus > or =7, an important predictor of final outcome.
Conclusions/significance: The method showed high sensitivity in a collection of specimens in which a significant portion of the total (13/31, equal to 42%) was considered CaP on the basis of having less than 15% of cancer cells. This result supports the notion of the "cancer field effect", in which transformed cells extend beyond morphologically evident tumour. The molecular diagnosis method here described is objective and less subjected to human error. Although further confirmations are needed, this method poses the potential to enhance conventional diagnosis.
Conflict of interest statement
Figures
References
-
- Pelzer AE, Tewari A, Bektic J, Berger AP, Frauscher, et al. Detection rates and biologic significance of prostate cancer with PSA less than 4.0 ng/mL: observation and clinical implications from Tyrol screening project. Urology. 2005;66:1029–33. - PubMed
-
- Huppi K, Chandramouli G. Molecular profiling of prostate cancer. Curr Urol Rep. 2004;5:45–51. - PubMed
-
- Bettuzzi S, Davalli P, Astancolle S, Carani C, Madeo B, et al. Tumor progression is accompanied by significant changes in the levels of expression of polyamine metabolism regulatory genes and Clusterin (Sulfated Glycoprotein 2) in human prostate cancer specimens. Cancer Res. 2000;60:28–34. - PubMed
-
- Bettuzzi S, Scaltriti M, Caporali A, Brausi M, D'Arca D, et al. Successful prediction of prostate cancer recurrence by gene profiling in combination with clinical data: a 5-year follow-up study. Cancer Res. 2003;63:3469–72. - PubMed
-
- Konishi H, Steinbach G, Terry N, Lee J, Dubin J, et al. Histone H3 messenger RNA in situ hybridization correlates with in vivo bromodeoxyuridine labeling of S-phase cells in rat colonic epithelium. Cancer Res. 1996;56:434–7. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

