Bridging the synaptic gap: neuroligins and neurexin I in Apis mellifera
- PMID: 18974885
- PMCID: PMC2570956
- DOI: 10.1371/journal.pone.0003542
Bridging the synaptic gap: neuroligins and neurexin I in Apis mellifera
Abstract
Vertebrate studies show neuroligins and neurexins are binding partners in a trans-synaptic cell adhesion complex, implicated in human autism and mental retardation disorders. Here we report a genetic analysis of homologous proteins in the honey bee. As in humans, the honeybee has five large (31-246 kb, up to 12 exons each) neuroligin genes, three of which are tightly clustered. RNA analysis of the neuroligin-3 gene reveals five alternatively spliced transcripts, generated through alternative use of exons encoding the cholinesterase-like domain. Whereas vertebrates have three neurexins the bee has just one gene named neurexin I (400 kb, 28 exons). However alternative isoforms of bee neurexin I are generated by differential use of 12 splice sites, mostly located in regions encoding LNS subdomains. Some of the splice variants of bee neurexin I resemble the vertebrate alpha- and beta-neurexins, albeit in vertebrates these forms are generated by alternative promoters. Novel splicing variations in the 3' region generate transcripts encoding alternative trans-membrane and PDZ domains. Another 3' splicing variation predicts soluble neurexin I isoforms. Neurexin I and neuroligin expression was found in brain tissue, with expression present throughout development, and in most cases significantly up-regulated in adults. Transcripts of neurexin I and one neuroligin tested were abundant in mushroom bodies, a higher order processing centre in the bee brain. We show neuroligins and neurexins comprise a highly conserved molecular system with likely similar functional roles in insects as vertebrates, and with scope in the honeybee to generate substantial functional diversity through alternative splicing. Our study provides important prerequisite data for using the bee as a model for vertebrate synaptic development.
Conflict of interest statement
Figures




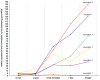
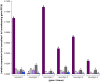


References
-
- Ichtchenko K, Hata Y, Nguyen T, Ullrich B, Missler M, et al. Neuroligin 1: a splice site-specific ligand for beta-neurexins. Cell. 1995;81:435–443. - PubMed
-
- Gilbert M, Smith J, Roskams AJ, Auld VJ. Neuroligin 3 is a vertebrate gliotactin expressed in the olfactory ensheathing glia, a growth-promoting class of macroglia. Glia. 2001;34:151–164. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

