The NIFSTD and BIRNLex vocabularies: building comprehensive ontologies for neuroscience
- PMID: 18975148
- PMCID: PMC2743139
- DOI: 10.1007/s12021-008-9032-z
The NIFSTD and BIRNLex vocabularies: building comprehensive ontologies for neuroscience
Abstract
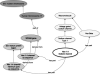
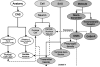
A critical component of the Neuroscience Information Framework (NIF) project is a consistent, flexible terminology for describing and retrieving neuroscience-relevant resources. Although the original NIF specification called for a loosely structured controlled vocabulary for describing neuroscience resources, as the NIF system evolved, the requirement for a formally structured ontology for neuroscience with sufficient granularity to describe and access a diverse collection of information became obvious. This requirement led to the NIF standardized (NIFSTD) ontology, a comprehensive collection of common neuroscience domain terminologies woven into an ontologically consistent, unified representation of the biomedical domains typically used to describe neuroscience data (e.g., anatomy, cell types, techniques), as well as digital resources (tools, databases) being created throughout the neuroscience community. NIFSTD builds upon a structure established by the BIRNLex, a lexicon of concepts covering clinical neuroimaging research developed by the Biomedical Informatics Research Network (BIRN) project. Each distinct domain module is represented using the Web Ontology Language (OWL). As much as has been practical, NIFSTD reuses existing community ontologies that cover the required biomedical domains, building the more specific concepts required to annotate NIF resources. By following this principle, an extensive vocabulary was assembled in a relatively short period of time for NIF information annotation, organization, and retrieval, in a form that promotes easy extension and modification. We report here on the structure of the NIFSTD, and its predecessor BIRNLex, the principles followed in its construction and provide examples of its use within NIF.
Figures





References
-
- Ascoli GA, Donohue DE, Halavi M. NeuroMorpho.Org: A central resource for neuronal morphologies. The Journal of Neuroscience. 2007;27:9247–9251. doi:10.1523/JNEUROSCI. 2055-07.2007. - PMC - PubMed
-
- Astakhov V, Gupta A, Grethe JS, Ross E, Little D, Yilmaz A, Ellisman M, et al. Semantically based data integration environment for biomedical research. Proc. of 19th IEEE Symp. on Comp. Based Med. Sys (CBMS'06) 2006
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

