A new method to compute K-mer frequencies and its application to annotate large repetitive plant genomes
- PMID: 18976482
- PMCID: PMC2613927
- DOI: 10.1186/1471-2164-9-517
A new method to compute K-mer frequencies and its application to annotate large repetitive plant genomes
Abstract
Background: The challenges of accurate gene prediction and enumeration are further aggravated in large genomes that contain highly repetitive transposable elements (TEs). Yet TEs play a substantial role in genome evolution and are themselves an important subject of study. Repeat annotation, based on counting occurrences of k-mers, has been previously used to distinguish TEs from low-copy genic regions; but currently available software solutions are impractical due to high memory requirements or specialization for specific user-tasks.
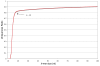
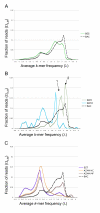
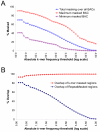
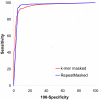
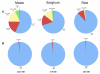
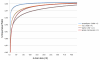
Results: Here we introduce the Tallymer software, a flexible and memory-efficient collection of programs for k-mer counting and indexing of large sequence sets. Unlike previous methods, Tallymer is based on enhanced suffix arrays. This gives a much larger flexibility concerning the choice of the k-mer size. Tallymer can process large data sizes of several billion bases. We used it in a variety of applications to study the genomes of maize and other plant species. In particular, Tallymer was used to index a set of whole genome shotgun sequences from maize (B73) (total size 109 bp.). We analyzed k-mer frequencies for a wide range of k. At this low genome coverage ( approximately 0.45x) highly repetitive 20-mers constituted 44% of the genome but represented only 1% of all possible k-mers. Similar low-complexity was seen in the repeat fractions of sorghum and rice. When applying our method to other maize data sets, High-C0t derived sequences showed the greatest enrichment for low-copy sequences. Among annotated TEs, the most highly repetitive were of the Ty3/gypsy class of retrotransposons, followed by the Ty1/copia class, and DNA transposons. Among expressed sequence tags (EST), a notable fraction contained high-copy k-mers, suggesting that transposons are still active in maize. Retrotransposons in Mo17 and McC cultivars were readily detected using the B73 20-mer frequency index, indicating their conservation despite extensive rearrangement across cultivars. Among one hundred annotated bacterial artificial chromosomes (BACs), k-mer frequency could be used to detect transposon-encoded genes with 92% sensitivity, compared to 96% using alignment-based repeat masking, while both methods showed 92% specificity.
Conclusion: The Tallymer software was effective in a variety of applications to aid genome annotation in maize, despite limitations imposed by the relatively low coverage of sequence available. For more information on the software, see http://www.zbh.uni-hamburg.de/Tallymer.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

