Head-head interaction characterizes the relaxed state of Limulus muscle myosin filaments
- PMID: 18976661
- PMCID: PMC4266557
- DOI: 10.1016/j.jmb.2008.10.038
Head-head interaction characterizes the relaxed state of Limulus muscle myosin filaments
Abstract
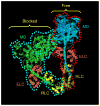
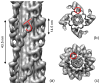
Regulation of muscle contraction via the myosin filaments occurs in vertebrate smooth and many invertebrate striated muscles. Studies of unphosphorylated vertebrate smooth muscle myosin suggest that activity is switched off through an intramolecular interaction between the actin-binding region of one head and the converter and essential light chains of the other, inhibiting ATPase activity and actin interaction. The same interaction (and additional interaction with the tail) is seen in three-dimensional reconstructions of relaxed, native myosin filaments from tarantula striated muscle, suggesting that such interactions are likely to underlie the off-state of myosin across a wide spectrum of the animal kingdom. We have tested this hypothesis by carrying out cryo-electron microscopy and three-dimensional image reconstruction of myosin filaments from horseshoe crab (Limulus) muscle. The same head-head and head-tail interactions seen in tarantula are also seen in Limulus, supporting the hypothesis. Other data suggest that this motif may underlie the relaxed state of myosin II in all species (including myosin II in nonmuscle cells), with the possible exception of insect flight muscle. The molecular organization of the myosin tails in the backbone of muscle thick filaments is unknown and may differ between species. X-ray diffraction data support a general model for crustaceans in which tails associate together to form 4-nm-diameter subfilaments, with these subfilaments assembling together to form the backbone. This model is supported by direct observation of 4-nm-diameter elongated strands in the tarantula reconstruction, suggesting that it might be a general structure across the arthropods. We observe a similar backbone organization in the Limulus reconstruction, supporting the general existence of such subfilaments.
Figures



References
-
- Ebashi S, Endo M, Ohtsuki I. Control of muscle contraction. Quart Rev Biophys. 1969;2:351–384. - PubMed
-
- Kendrick-Jones J, Lehman W, Szent-Gyorgyi AG. Regulation in molluscan muscles. J Mol Biol. 1970;54:313–326. - PubMed
-
- Szent-Gyorgyi AG, Kalabokis VN, Perreault-Micale CL. Regulation by molluscan myosins. Mol Cell Biochem. 1999;190:55–62. - PubMed
-
- Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80:853–924. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

