Detection of protein S-nitrosylation with the biotin-switch technique
- PMID: 18977293
- PMCID: PMC3120222
- DOI: 10.1016/j.freeradbiomed.2008.09.034
Detection of protein S-nitrosylation with the biotin-switch technique
Abstract
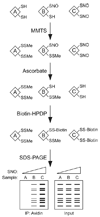
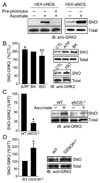
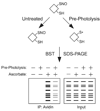
Protein S-nitrosylation, the posttranslational modification of cysteine thiols to form S-nitrosothiols, is a principle mechanism of nitric oxide-based signaling. Studies have demonstrated myriad roles for S-nitrosylation in organisms from bacteria to humans, and recent efforts have greatly advanced our scientific understanding of how this redox-based modification is dynamically regulated during physiological and pathophysiological conditions. The focus of this review is the biotin-switch technique (BST), which has become a mainstay assay for detecting S-nitrosylated proteins in complex biological systems. Potential pitfalls and modern adaptations of the BST are discussed, as are future directions for this assay in the burgeoning field of protein S-nitrosylation.
Figures


Similar articles
-
Analysis of Recombinant Protein S-Nitrosylation Using the Biotin-Switch Technique.Methods Mol Biol. 2018;1747:131-141. doi: 10.1007/978-1-4939-7695-9_11. Methods Mol Biol. 2018. PMID: 29600456
-
Assessment and application of the biotin switch technique for examining protein S-nitrosylation under conditions of pharmacologically induced oxidative stress.J Biol Chem. 2007 May 11;282(19):13977-83. doi: 10.1074/jbc.M609684200. Epub 2007 Mar 21. J Biol Chem. 2007. PMID: 17376775
-
Sinapinic acid can replace ascorbate in the biotin switch assay.Biochim Biophys Acta. 2010 Jan;1800(1):23-30. doi: 10.1016/j.bbagen.2009.10.004. Epub 2009 Oct 23. Biochim Biophys Acta. 2010. PMID: 19837133
-
Protein s-nitrosylation measurement.Methods Enzymol. 2013;522:409-25. doi: 10.1016/B978-0-12-407865-9.00019-4. Methods Enzymol. 2013. PMID: 23374195 Review.
-
Screening systems for the identification of S-nitrosylated proteins.Nitric Oxide. 2011 Aug 1;25(2):108-11. doi: 10.1016/j.niox.2010.11.002. Epub 2010 Nov 24. Nitric Oxide. 2011. PMID: 21111056 Review.
Cited by
-
Proteomic approaches to quantify cysteine reversible modifications in aging and neurodegenerative diseases.Proteomics Clin Appl. 2016 Dec;10(12):1159-1177. doi: 10.1002/prca.201600015. Epub 2016 Nov 11. Proteomics Clin Appl. 2016. PMID: 27666938 Free PMC article. Review.
-
Effect of Nitrosative Stress on the S-Nitroso-Proteome of Paracoccidioides brasiliensis.Front Microbiol. 2020 Jun 4;11:1184. doi: 10.3389/fmicb.2020.01184. eCollection 2020. Front Microbiol. 2020. PMID: 32582109 Free PMC article.
-
Potential widespread denitrosylation of brain proteins following prolonged restraint: proposed links between stress and central nervous system disease.Metab Brain Dis. 2019 Feb;34(1):183-189. doi: 10.1007/s11011-018-0340-1. Epub 2018 Nov 9. Metab Brain Dis. 2019. PMID: 30414012
-
Cytosolic Isocitrate Dehydrogenase from Arabidopsis thaliana Is Regulated by Glutathionylation.Antioxidants (Basel). 2019 Jan 8;8(1):16. doi: 10.3390/antiox8010016. Antioxidants (Basel). 2019. PMID: 30625997 Free PMC article.
-
Opening of Cx43-formed hemichannels mediates the Ca2+ signaling associated with endothelial cell migration.Biol Direct. 2023 Aug 28;18(1):52. doi: 10.1186/s13062-023-00408-3. Biol Direct. 2023. PMID: 37635249 Free PMC article.
References
-
- Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. - PubMed
-
- Foster MW, McMahon TJ, Stamler JS. S-nitrosylation in health and disease. Trends Mol Med. 2003;9:160–168. - PubMed
-
- Nozik-Grayck E, Whalen EJ, Stamler JS, McMahon TJ, Chitano P, Piantadosi CA. S-nitrosoglutathione inhibits alpha1-adrenergic receptor-mediated vasoconstriction and ligand binding in pulmonary artery. Am J Physiol Lung Cell Mol Physiol. 2006;290:L136–L143. - PubMed
-
- Whalen EJ, Foster MW, Matsumoto A, Ozawa K, Violin JD, Que LG, Nelson CD, Benhar M, Keys JR, Rockman HA, Koch WJ, Daaka Y, Lefkowitz RJ, Stamler JS. Regulation of beta-adrenergic receptor signaling by S-nitrosylation of G-protein-coupled receptor kinase 2. Cell. 2007;129:511–522. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

