H3K79 methylation profiles define murine and human MLL-AF4 leukemias
- PMID: 18977325
- PMCID: PMC2591932
- DOI: 10.1016/j.ccr.2008.10.001
H3K79 methylation profiles define murine and human MLL-AF4 leukemias
Abstract
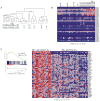
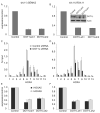
We created a mouse model wherein conditional expression of an Mll-AF4 fusion oncogene induces B precursor acute lymphoblastic (ALL) or acute myeloid leukemias (AML). Gene expression profile analysis of the ALL cells demonstrated significant overlap with human MLL-rearranged ALL. ChIP-chip analysis demonstrated histone H3 lysine 79 (H3K79) methylation profiles that correlated with Mll-AF4-associated gene expression profiles in murine ALLs and in human MLL-rearranged leukemias. Human MLL-rearranged ALLs could be distinguished from other ALLs by their H3K79 profiles, and suppression of the H3K79 methyltransferase DOT1L inhibited expression of critical MLL-AF4 target genes. We thus demonstrate that ectopic H3K79 methylation is a distinguishing feature of murine and human MLL-AF4 ALLs and is important for maintenance of MLL-AF4-driven gene expression.
Figures





Comment in
-
Reconstructing the disease model and epigenetic networks for MLL-AF4 leukemia.Cancer Cell. 2008 Nov 4;14(5):345-7. doi: 10.1016/j.ccr.2008.10.008. Cancer Cell. 2008. PMID: 18977321
References
-
- Armstrong SA, Kung AL, Mabon ME, Silverman LB, Stam RW, Den Boer ML, Pieters R, Kersey JH, Sallan SE, Fletcher JA, et al. Inhibition of FLT3 in MLL. Validation of a therapeutic target identified by gene expression based classification. Cancer Cell. 2003;3:173–183. - PubMed
-
- Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. - PubMed
-
- Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20:5695–5707. - PubMed
-
- Biondi A, Cimino G, Pieters R, Pui CH. Biological and therapeutic aspects of infant leukemia. Blood. 2000;96:24–33. - PubMed
-
- Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

