Petabyte-scale innovations at the European Nucleotide Archive
- PMID: 18978013
- PMCID: PMC2686451
- DOI: 10.1093/nar/gkn765
Petabyte-scale innovations at the European Nucleotide Archive
Abstract
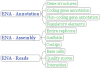
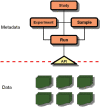
Dramatic increases in the throughput of nucleotide sequencing machines, and the promise of ever greater performance, have thrust bioinformatics into the era of petabyte-scale data sets. Sequence repositories, which provide the feed for these data sets into the worldwide computational infrastructure, are challenged by the impact of these data volumes. The European Nucleotide Archive (ENA; http://www.ebi.ac.uk/embl), comprising the EMBL Nucleotide Sequence Database and the Ensembl Trace Archive, has identified challenges in the storage, movement, analysis, interpretation and visualization of petabyte-scale data sets. We present here our new repository for next generation sequence data, a brief summary of contents of the ENA and provide details of major developments to submission pipelines, high-throughput rule-based validation infrastructure and data integration approaches.
Figures