ClustScan: an integrated program package for the semi-automatic annotation of modular biosynthetic gene clusters and in silico prediction of novel chemical structures
- PMID: 18978015
- PMCID: PMC2588505
- DOI: 10.1093/nar/gkn685
ClustScan: an integrated program package for the semi-automatic annotation of modular biosynthetic gene clusters and in silico prediction of novel chemical structures
Abstract
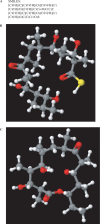
The program package 'ClustScan' (Cluster Scanner) is designed for rapid, semi-automatic, annotation of DNA sequences encoding modular biosynthetic enzymes including polyketide synthases (PKS), non-ribosomal peptide synthetases (NRPS) and hybrid (PKS/NRPS) enzymes. The program displays the predicted chemical structures of products as well as allowing export of the structures in a standard format for analyses with other programs. Recent advances in understanding of enzyme function are incorporated to make knowledge-based predictions about the stereochemistry of products. The program structure allows easy incorporation of additional knowledge about domain specificities and function. The results of analyses are presented to the user in a graphical interface, which also allows easy editing of the predictions to incorporate user experience. The versatility of this program package has been demonstrated by annotating biochemical pathways in microbial, invertebrate animal and metagenomic datasets. The speed and convenience of the package allows the annotation of all PKS and NRPS clusters in a complete Actinobacteria genome in 2-3 man hours. The open architecture of ClustScan allows easy integration with other programs, facilitating further analyses of results, which is useful for a broad range of researchers in the chemical and biological sciences.
Figures


References
-
- Challis GL. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chembiochem. 2005;6:601–611. - PubMed
-
- Finking R, Marahiel MA. Biosynthesis of non-ribosomal peptides. Ann. Rev. Microbiol. 2004;58:453–488. - PubMed
-
- Hranueli D, Cullum J, Basrak B, Goldstein P, Long PF. Plasticity of the Streptomyces genome - evolution and engineering of new antibiotics. Curr. Med. Chem. 2005;12:1697–1704. - PubMed
-
- Weissman KJ, Leadlay PF. Combinatorial biosynthesis of reduced polyketides. Nat. Rev. Microbiol. 2005;3:925–936. - PubMed
-
- Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

