EFD Is an ERF transcription factor involved in the control of nodule number and differentiation in Medicago truncatula
- PMID: 18978033
- PMCID: PMC2590733
- DOI: 10.1105/tpc.108.059857
EFD Is an ERF transcription factor involved in the control of nodule number and differentiation in Medicago truncatula
Abstract
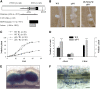
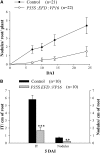
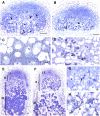
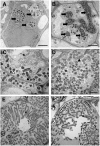
Mechanisms regulating legume root nodule development are still poorly understood, and very few regulatory genes have been cloned and characterized. Here, we describe EFD (for ethylene response factor required for nodule differentiation), a gene that is upregulated during nodulation in Medicago truncatula. The EFD transcription factor belongs to the ethylene response factor (ERF) group V, which contains ERN1, 2, and 3, three ERFs involved in Nod factor signaling. The role of EFD in the regulation of nodulation was examined through the characterization of a null deletion mutant (efd-1), RNA interference, and overexpression studies. These studies revealed that EFD is a negative regulator of root nodulation and infection by Rhizobium and that EFD is required for the formation of functional nitrogen-fixing nodules. EFD appears to be involved in the plant and bacteroid differentiation processes taking place beneath the nodule meristem. We also showed that EFD activated Mt RR4, a cytokinin primary response gene that encodes a type-A response regulator. We propose that EFD induction of Mt RR4 leads to the inhibition of cytokinin signaling, with two consequences: the suppression of new nodule initiation and the activation of differentiation as cells leave the nodule meristem. Our work thus reveals a key regulator linking early and late stages of nodulation and suggests that the regulation of the cytokinin pathway is important both for nodule initiation and development.
Figures


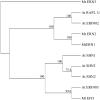

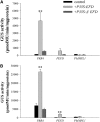
References
-
- Alunni, B., Kevei, Z., Redondo-Nieto, M., Kondorosi, A., Mergaert, P., and Kondorosi, E. (2007). Genomic organization and evolutionary insights on GRP and NCR genes, two large nodule-specific gene families in Medicago truncatula. Mol. Plant Microbe Interact. 20 1138–1148. - PubMed
-
- Ardourel, M., Demont, N., Debelle, F., Maillet, F., de Billy, F., Prome, J.C., Denarie, J., and Truchet, G. (1994). Rhizobium meliloti lipooligosaccharide nodulation factors: Different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6 1357–1374. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

