A genomic distance based on MUM indicates discontinuity between most bacterial species and genera
- PMID: 18978054
- PMCID: PMC2612450
- DOI: 10.1128/JB.01202-08
A genomic distance based on MUM indicates discontinuity between most bacterial species and genera
Abstract
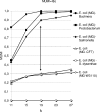
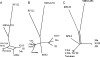
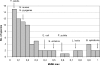
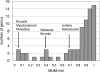
The fundamental unit of biological diversity is the species. However, a remarkable extent of intraspecies diversity in bacteria was discovered by genome sequencing, and it reveals the need to develop clear criteria to group strains within a species. Two main types of analyses used to quantify intraspecies variation at the genome level are the average nucleotide identity (ANI), which detects the DNA conservation of the core genome, and the DNA content, which calculates the proportion of DNA shared by two genomes. Both estimates are based on BLAST alignments for the definition of DNA sequences common to the genome pair. Interestingly, however, results using these methods on intraspecies pairs are not well correlated. This prompted us to develop a genomic-distance index taking into account both criteria of diversity, which are based on DNA maximal unique matches (MUM) shared by two genomes. The values, called MUMi, for MUM index, correlate better with the ANI than with the DNA content. Moreover, the MUMi groups strains in a way that is congruent with routinely used multilocus sequence-typing trees, as well as with ANI-based trees. We used the MUMi to determine the relatedness of all available genome pairs at the species and genus levels. Our analysis reveals a certain consistency in the current notion of bacterial species, in that the bulk of intraspecies and intragenus values are clearly separable. It also confirms that some species are much more diverse than most. As the MUMi is fast to calculate, it offers the possibility of measuring genome distances on the whole database of available genomes.
Figures


Similar articles
-
Toward a more robust assessment of intraspecies diversity, using fewer genetic markers.Appl Environ Microbiol. 2006 Nov;72(11):7286-93. doi: 10.1128/AEM.01398-06. Epub 2006 Sep 15. Appl Environ Microbiol. 2006. PMID: 16980418 Free PMC article.
-
Genomic insights that advance the species definition for prokaryotes.Proc Natl Acad Sci U S A. 2005 Feb 15;102(7):2567-72. doi: 10.1073/pnas.0409727102. Epub 2005 Feb 8. Proc Natl Acad Sci U S A. 2005. PMID: 15701695 Free PMC article.
-
ANItools web: a web tool for fast genome comparison within multiple bacterial strains.Database (Oxford). 2016 Jun 5;2016:baw084. doi: 10.1093/database/baw084. Print 2016. Database (Oxford). 2016. PMID: 27270714 Free PMC article.
-
Bacterial strain typing in the genomic era.FEMS Microbiol Rev. 2009 Sep;33(5):892-916. doi: 10.1111/j.1574-6976.2009.00182.x. Epub 2009 May 6. FEMS Microbiol Rev. 2009. PMID: 19453749 Review.
-
The diverse and dynamic structure of bacterial genomes.Annu Rev Genet. 1998;32:339-77. doi: 10.1146/annurev.genet.32.1.339. Annu Rev Genet. 1998. PMID: 9928484 Review.
Cited by
-
Shifting the genomic gold standard for the prokaryotic species definition.Proc Natl Acad Sci U S A. 2009 Nov 10;106(45):19126-31. doi: 10.1073/pnas.0906412106. Epub 2009 Oct 23. Proc Natl Acad Sci U S A. 2009. PMID: 19855009 Free PMC article.
-
Ralstonia solanacearum requires PopS, an ancient AvrE-family effector, for virulence and To overcome salicylic acid-mediated defenses during tomato pathogenesis.mBio. 2013 Nov 26;4(6):e00875-13. doi: 10.1128/mBio.00875-13. mBio. 2013. PMID: 24281716 Free PMC article.
-
Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison.Stand Genomic Sci. 2010 Jan 28;2(1):117-34. doi: 10.4056/sigs.531120. Stand Genomic Sci. 2010. PMID: 21304684 Free PMC article.
-
Genomic Comparisons Reveal Microevolutionary Differences in Mycobacterium abscessus Subspecies.Front Microbiol. 2017 Oct 23;8:2042. doi: 10.3389/fmicb.2017.02042. eCollection 2017. Front Microbiol. 2017. PMID: 29109707 Free PMC article.
-
Comparative genomics of 12 strains of Erwinia amylovora identifies a pan-genome with a large conserved core.PLoS One. 2013;8(2):e55644. doi: 10.1371/journal.pone.0055644. Epub 2013 Feb 7. PLoS One. 2013. PMID: 23409014 Free PMC article.
References
-
- Canchaya, C., M. J. Claesson, G. F. Fitzgerald, D. van Sinderen, and P. W. O'Toole. 2006. Diversity of the genus Lactobacillus revealed by comparative genomics of five species. Microbiology 1523185-3196. - PubMed
-
- Chain, P. S., E. Carniel, F. W. Larimer, J. Lamerdin, P. O. Stoutland, W. M. Regala, A. M. Georgescu, L. M. Vergez, M. L. Land, V. L. Motin, R. R. Brubaker, J. Fowler, J. Hinnebusch, M. Marceau, C. Medigue, M. Simonet, V. Chenal-Francisque, B. Souza, D. Dacheux, J. M. Elliott, A. Derbise, L. J. Hauser, and E. Garcia. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 10113826-31. - PMC - PubMed
-
- Chen, X., M. Li, B. Ma, and J. Tromp. 2002. DNACompress: fast and effective DNA sequence compression. Bioinformatics 181696-1698. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

