Streptococcus mutans SMU.623c codes for a functional, metal-dependent polysaccharide deacetylase that modulates interactions with salivary agglutinin
- PMID: 18978064
- PMCID: PMC2612446
- DOI: 10.1128/JB.00838-08
Streptococcus mutans SMU.623c codes for a functional, metal-dependent polysaccharide deacetylase that modulates interactions with salivary agglutinin
Abstract
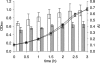
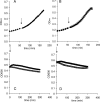
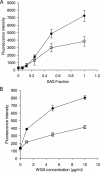
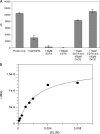
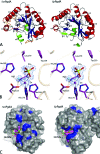
The genome sequence of the oral pathogen Streptococcus mutans predicts the presence of two putative polysaccharide deacetylases. The first, designated PgdA in this paper, shows homology to the catalytic domains of peptidoglycan deacetylases from Streptococcus pneumoniae and Listeria monocytogenes, which are both thought to be involved in the bacterial defense mechanism against human mucosal lysozyme and are part of the CAZY family 4 carbohydrate esterases. S. mutans cells in which the pgdA gene was deleted displayed a different colony texture and a slightly increased cell surface hydrophobicity and yet did not become hypersensitive to lysozyme as shown previously for S. pneumoniae. To understand this apparent lack of activity, the high-resolution X-ray structure of S. mutans PgdA was determined; it showed the typical carbohydrate esterase 4 fold, with metal bound in a His-His-Asp triad. Analysis of the protein surface showed that an extended groove lined with aromatic residues is orientated toward the active-site residues. The protein exhibited metal-dependent de-N-acetylase activity toward a hexamer of N-acetylglucosamine. No activity was observed toward shorter chitooligosaccharides or a synthetic peptidoglycan tetrasaccharide. In agreement with the lysozyme data this would suggest that S. mutans PgdA does not act on peptidoglycan but on an as-yet-unidentified polysaccharide within the bacterial cell surface. Strikingly, the pgdA-knockout strain showed a significant increase in aggregation/agglutination by salivary agglutinin, in agreement with this gene acting as a deacetylase of a cell surface glycan.
Figures


Similar articles
-
The pgdA gene encodes for a peptidoglycan N-acetylglucosamine deacetylase in Streptococcus pneumoniae.J Biol Chem. 2000 Jul 7;275(27):20496-501. doi: 10.1074/jbc.M910189199. J Biol Chem. 2000. PMID: 10781617
-
Inactivation of the wall-associated de-N-acetylase (PgdA) of Listeria monocytogenes results in greater susceptibility of the cells to induced autolysis.J Microbiol Biotechnol. 2009 Sep;19(9):932-45. doi: 10.4014/jmb.0810.557. J Microbiol Biotechnol. 2009. PMID: 19809250
-
Lysozyme Resistance in Clostridioides difficile Is Dependent on Two Peptidoglycan Deacetylases.J Bacteriol. 2020 Oct 22;202(22):e00421-20. doi: 10.1128/JB.00421-20. Print 2020 Oct 22. J Bacteriol. 2020. PMID: 32868404 Free PMC article.
-
Characterization of a P1-deficient strain of Streptococcus mutans that expresses the SpaA protein of Streptococcus sobrinus.Infect Immun. 1996 Sep;64(9):3652-8. doi: 10.1128/iai.64.9.3652-3658.1996. Infect Immun. 1996. PMID: 8751913 Free PMC article.
-
Resistance to mucosal lysozyme compensates for the fitness deficit of peptidoglycan modifications by Streptococcus pneumoniae.PLoS Pathog. 2008 Dec;4(12):e1000241. doi: 10.1371/journal.ppat.1000241. Epub 2008 Dec 12. PLoS Pathog. 2008. PMID: 19079576 Free PMC article.
Cited by
-
Development of a fluorescence-based excipient screening for improved stability and shelf-life of recombinant chitin deacetylase.Biochem Biophys Rep. 2024 Apr 27;38:101718. doi: 10.1016/j.bbrep.2024.101718. eCollection 2024 Jul. Biochem Biophys Rep. 2024. PMID: 38708424 Free PMC article.
-
Two Putative Polysaccharide Deacetylases Are Required for Osmotic Stability and Cell Shape Maintenance in Bacillus anthracis.J Biol Chem. 2015 May 22;290(21):13465-78. doi: 10.1074/jbc.M115.640029. Epub 2015 Mar 30. J Biol Chem. 2015. PMID: 25825488 Free PMC article.
-
The Cell Wall Deacetylases Spy1094 and Spy1370 Contribute to Streptococcus pyogenes Virulence.Microorganisms. 2023 Jan 24;11(2):305. doi: 10.3390/microorganisms11020305. Microorganisms. 2023. PMID: 36838272 Free PMC article.
-
Substrate Recognition and Specificity of Chitin Deacetylases and Related Family 4 Carbohydrate Esterases.Int J Mol Sci. 2018 Jan 30;19(2):412. doi: 10.3390/ijms19020412. Int J Mol Sci. 2018. PMID: 29385775 Free PMC article. Review.
-
Crystal structure of acetylxylan esterase from Caldanaerobacter subterraneus subsp. tengcongensis.Acta Crystallogr F Struct Biol Commun. 2021 Nov 1;77(Pt 11):399-406. doi: 10.1107/S2053230X21009675. Epub 2021 Oct 19. Acta Crystallogr F Struct Biol Commun. 2021. PMID: 34726178 Free PMC article.
References
-
- Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 9914434-14439. - PMC - PubMed
-
- Bikker, F. J., A. J. Ligtenberg, C. End, M. Renner, S. Blaich, S. Lyer, R. Wittig, W. van't Hof, E. C. Veerman, K. Nazmi, J. M. de Blieck-Hogervorst, P. Kioschis, A. V. Nieuw Amerongen, A. Poustka, and J. Mollenhauer. 2004. Bacteria binding by DMBT1/SAG/gp-340 is confined to the VEVLXXXXW motif in its scavenger receptor cysteine-rich domains. J. Biol. Chem. 27947699-47703. - PubMed
-
- Bikker, F. J., A. J. Ligtenberg, K. Nazmi, E. C. Veerman, W. van't Hof, J. G. Bolscher, A. Poustka, A. V. Nieuw Amerongen, and J. Mollenhauer. 2002. Identification of the bacteria-binding peptide domain on salivary agglutinin (gp-340/DMBT1), a member of the scavenger receptor cysteine-rich superfamily. J. Biol. Chem. 27732109-32115. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

