Streptococcus mutans SMU.623c codes for a functional, metal-dependent polysaccharide deacetylase that modulates interactions with salivary agglutinin
- PMID: 18978064
- PMCID: PMC2612446
- DOI: 10.1128/JB.00838-08
Streptococcus mutans SMU.623c codes for a functional, metal-dependent polysaccharide deacetylase that modulates interactions with salivary agglutinin
Abstract
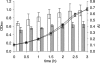
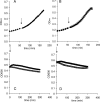
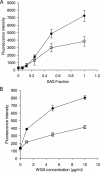
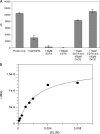
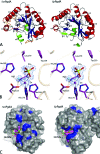
The genome sequence of the oral pathogen Streptococcus mutans predicts the presence of two putative polysaccharide deacetylases. The first, designated PgdA in this paper, shows homology to the catalytic domains of peptidoglycan deacetylases from Streptococcus pneumoniae and Listeria monocytogenes, which are both thought to be involved in the bacterial defense mechanism against human mucosal lysozyme and are part of the CAZY family 4 carbohydrate esterases. S. mutans cells in which the pgdA gene was deleted displayed a different colony texture and a slightly increased cell surface hydrophobicity and yet did not become hypersensitive to lysozyme as shown previously for S. pneumoniae. To understand this apparent lack of activity, the high-resolution X-ray structure of S. mutans PgdA was determined; it showed the typical carbohydrate esterase 4 fold, with metal bound in a His-His-Asp triad. Analysis of the protein surface showed that an extended groove lined with aromatic residues is orientated toward the active-site residues. The protein exhibited metal-dependent de-N-acetylase activity toward a hexamer of N-acetylglucosamine. No activity was observed toward shorter chitooligosaccharides or a synthetic peptidoglycan tetrasaccharide. In agreement with the lysozyme data this would suggest that S. mutans PgdA does not act on peptidoglycan but on an as-yet-unidentified polysaccharide within the bacterial cell surface. Strikingly, the pgdA-knockout strain showed a significant increase in aggregation/agglutination by salivary agglutinin, in agreement with this gene acting as a deacetylase of a cell surface glycan.
Figures


References
-
- Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 9914434-14439. - PMC - PubMed
-
- Bikker, F. J., A. J. Ligtenberg, C. End, M. Renner, S. Blaich, S. Lyer, R. Wittig, W. van't Hof, E. C. Veerman, K. Nazmi, J. M. de Blieck-Hogervorst, P. Kioschis, A. V. Nieuw Amerongen, A. Poustka, and J. Mollenhauer. 2004. Bacteria binding by DMBT1/SAG/gp-340 is confined to the VEVLXXXXW motif in its scavenger receptor cysteine-rich domains. J. Biol. Chem. 27947699-47703. - PubMed
-
- Bikker, F. J., A. J. Ligtenberg, K. Nazmi, E. C. Veerman, W. van't Hof, J. G. Bolscher, A. Poustka, A. V. Nieuw Amerongen, and J. Mollenhauer. 2002. Identification of the bacteria-binding peptide domain on salivary agglutinin (gp-340/DMBT1), a member of the scavenger receptor cysteine-rich superfamily. J. Biol. Chem. 27732109-32115. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

