FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts
- PMID: 18978354
- PMCID: PMC2662291
- DOI: 10.1074/jbc.M808152200
FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts
Abstract
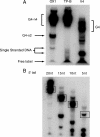
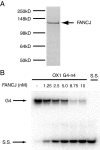
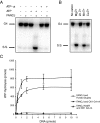
Fanconi anemia (FA) is a heritable human cancer-susceptibility disorder, delineating a genetically heterogenous pathway for the repair of replication-blocking lesions such as interstrand DNA cross-links. Here we demonstrate that one component of this pathway, FANCJ, is a structure-specific DNA helicase that dissociates guanine quadruplex DNA (G4 DNA) in vitro. Moreover, in contrast with previously identified G4 DNA helicases, such as the Bloom's helicase (BLM), FANCJ unwinds G4 substrates with 5'-3' polarity. In the FA-J human patient cell line EUFA0030 the loss of FANCJ G4 unwinding function correlates with the accumulation of large genomic deletions in the vicinity of sequences, which match the G4 DNA signature. Together these findings support a role for FANCJ in the maintenance of potentially unstable genomic G/C tracts during replication.
Figures




References
-
- Seal, S., Thompson, D., Renwick, A., Elliott, A., Kelly, P., Barfoot, R., Chagtai, T., Jayatilake, H., Ahmed, M., Spanova, K., North, B., McGuffog, L., Evans, D. G., Eccles, D., Easton, D. F., Stratton, M. R., and Rahman, N. (2006) Nat. Genet. 38 1239–1241 - PubMed
-
- Levitus, M., Waisfisz, Q., Godthelp, B. C., de Vries, Y., Hussain, S., Wiegant, W. W., Elghalbzouri-Maghrani, E., Steltenpool, J., Rooimans, M. A., Pals, G., Arwert, F., Mathew, C. G., Zdzienicka, M. Z., Hiom, K., De Winter, J. P., and Joenje, H. (2005) Nat. Genet. 37 934–935 - PubMed
-
- Litman, R., Peng, M., Jin, Z., Zhang, F., Zhang, J., Powell, S., Andreassen, P. R., and Cantor, S. B. (2005) Cancer Cell 8 255–265 - PubMed
-
- Levran, O., Attwooll, C., Henry, R. T., Milton, K. L., Neveling, K., Rio, P., Batish, S. D., Kalb, R., Velleuer, E., Barral, S., Ott, J., Petrini, J., Schindler, D., Hanenberg, H., and Auerbach, A. D. (2005) Nat. Genet. 37 931–933 - PubMed
-
- Joenje, H., and Patel, K. J. (2001) Nat. Rev. 2 446–457 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

