Modification of quinone electrochemistry by the proteins in the biological electron transfer chains: examples from photosynthetic reaction centers
- PMID: 18979192
- PMCID: PMC2746407
- DOI: 10.1007/s10863-008-9179-1
Modification of quinone electrochemistry by the proteins in the biological electron transfer chains: examples from photosynthetic reaction centers
Abstract
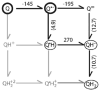
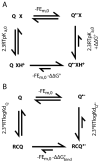
Quinones such as ubiquinone are the lipid soluble electron and proton carriers in the membranes of mitochondria, chloroplasts and oxygenic bacteria. Quinones undergo controlled redox reactions bound to specific sites in integral membrane proteins such as the cytochrome bc(1) oxidoreductase. The quinone reactions in bacterial photosynthesis are amongst the best characterized, presenting a model to understand how proteins modulate cofactor chemistry. The free energy of ubiquinone redox reactions in aqueous solution and in the Q(A) and Q(B) sites of the bacterial photosynthetic reaction centers (RCs) are compared. In the primary Q(A) site ubiquinone is reduced only to the anionic semiquinone (Q(*-)) while in the secondary Q(B) site the product is the doubly reduced, doubly protonated quinol (QH(2)). The ways in which the protein modifies the relative energy of each reduced and protonated intermediate are described. For example, the protein stabilizes Q(*-) while destabilizing Q(=) relative to aqueous solution through electrostatic interactions. In addition, kinetic and thermodynamic mechanisms for stabilizing the intermediate semiquinones are compared. Evidence for the protein sequestering anionic compounds by slowing both on and off rates as well as by binding the anion more tightly is reviewed.
Figures
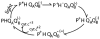

Similar articles
-
Slow dissociation of a charged ligand: analysis of the primary quinone Q(A) site of photosynthetic bacterial reaction centers.J Am Chem Soc. 2011 Nov 2;133(43):17375-85. doi: 10.1021/ja205811f. Epub 2011 Oct 11. J Am Chem Soc. 2011. PMID: 21863833 Free PMC article.
-
Energetics of quinone-dependent electron and proton transfers in Rhodobacter sphaeroides photosynthetic reaction centers.Biochemistry. 2005 Jan 11;44(1):82-96. doi: 10.1021/bi048348k. Biochemistry. 2005. PMID: 15628848
-
Redox potential tuning through differential quinone binding in the photosynthetic reaction center of Rhodobacter sphaeroides.Biochemistry. 2015 Mar 31;54(12):2104-16. doi: 10.1021/acs.biochem.5b00033. Epub 2015 Mar 23. Biochemistry. 2015. PMID: 25734689 Free PMC article.
-
The Q cycle of cytochrome bc complexes: a structure perspective.Biochim Biophys Acta. 2011 Jul;1807(7):788-802. doi: 10.1016/j.bbabio.2011.02.006. Epub 2011 Feb 23. Biochim Biophys Acta. 2011. PMID: 21352799 Free PMC article. Review.
-
Coupling of electron transfer to proton uptake at the Q(B) site of the bacterial reaction center: a perspective from FTIR difference spectroscopy.Biochim Biophys Acta. 2008 Oct;1777(10):1229-48. doi: 10.1016/j.bbabio.2008.06.012. Epub 2008 Jul 11. Biochim Biophys Acta. 2008. PMID: 18671937 Review.
Cited by
-
Energetics of the exchangeable quinone, QB, in Photosystem II.Proc Natl Acad Sci U S A. 2019 Sep 24;116(39):19458-19463. doi: 10.1073/pnas.1910675116. Epub 2019 Sep 5. Proc Natl Acad Sci U S A. 2019. PMID: 31488720 Free PMC article.
-
Tools for analyzing protonation states and for tracing proton transfer pathways with examples from the Rb. sphaeroides photosynthetic reaction centers.Photosynth Res. 2023 Apr;156(1):101-112. doi: 10.1007/s11120-022-00973-0. Epub 2022 Oct 29. Photosynth Res. 2023. PMID: 36307598
-
Exploring by pulsed EPR the electronic structure of ubisemiquinone bound at the QH site of cytochrome bo3 from Escherichia coli with in vivo 13C-labeled methyl and methoxy substituents.J Biol Chem. 2011 Mar 25;286(12):10105-14. doi: 10.1074/jbc.M110.206821. Epub 2011 Jan 19. J Biol Chem. 2011. PMID: 21247900 Free PMC article.
-
Distinct properties of semiquinone species detected at the ubiquinol oxidation Qo site of cytochrome bc1 and their mechanistic implications.J R Soc Interface. 2016 May;13(118):20160133. doi: 10.1098/rsif.2016.0133. J R Soc Interface. 2016. PMID: 27194483 Free PMC article. Review.
-
Comparison of proton transfer paths to the QA and QB sites of the Rb. sphaeroides photosynthetic reaction centers.Photosynth Res. 2022 May;152(2):153-165. doi: 10.1007/s11120-022-00906-x. Epub 2022 Mar 28. Photosynth Res. 2022. PMID: 35344134
References
-
- Alexov EG, Gunner MR. Calculated protein and proton motions coupled to electron transfer: electron transfer from QA− to QB in bacterial photosynthetic reaction centers. Biochemistry. 1999;38:8253–8270. - PubMed
-
- Alexov E, Miksovska J, Baciou L, Schiffer M, Hanson DK, Sebban P, Gunner MR. Modeling the effects of mutations on the free energy of the first electron transfer from QA− to QB in photosynthetic reaction centers. Biochemistry. 2000;39:5940–5952. - PubMed
-
- Baker NA. Improving implicit solvent simulations: a Poisson-centric view. Cur Opin Struct Biol. 2005;15:137–143. - PubMed
-
- Bashford D, Karplus M. The pKas of ionizable groups in proteins: atomic detail from a continuum electrostatic model. Biochemistry. 1990;29:10219–10225. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

