Intestinal macrophage/epithelial cell-derived CCL11/eotaxin-1 mediates eosinophil recruitment and function in pediatric ulcerative colitis
- PMID: 18981162
- PMCID: PMC2728352
- DOI: 10.4049/jimmunol.181.10.7390
Intestinal macrophage/epithelial cell-derived CCL11/eotaxin-1 mediates eosinophil recruitment and function in pediatric ulcerative colitis
Abstract
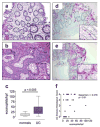
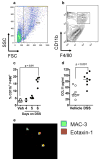
Clinical studies have demonstrated a link between the eosinophil-selective chemokines, eotaxins (eotaxin-1/CCL11 and eotaxin-2/CCL24), eosinophils, and the inflammatory bowel diseases, Crohn's disease and ulcerative colitis (UC). However, the cellular source and individual contribution of the eotaxins to colonic eosinophilic accumulation in inflammatory bowel diseases remain unclear. In this study we demonstrate, by gene array and quantitative PCR, elevated levels of eotaxin-1 mRNA in the rectosigmoid colon of pediatric UC patients. We show that elevated levels of eotaxin-1 mRNA positively correlated with rectosigmoid eosinophil numbers. Further, colonic eosinophils appeared to be degranulating, and the levels positively correlated with disease severity. Using the dextran sodium sulfate (DSS)-induced intestinal epithelial injury model, we show that DSS treatment of mice strongly induced colonic eotaxin-1 and eotaxin-2 expression and eosinophil levels. Analysis of eosinophil-deficient mice defined an effector role for eosinophils in disease pathology. DSS treatment of eotaxin-2(-/-) and eotaxin-1/2(-/-) mice demonstrated that eosinophil recruitment was dependent on eotaxin-1. In situ and immunofluorescence analysis-identified eotaxin-1 expression was restricted to intestinal F4/80(+)CD11b(+) macrophages in DSS-induced epithelial injury and to CD68(+) intestinal macrophages and the basolateral compartment of intestinal epithelial cells in pediatric UC. These data demonstrate that intestinal macrophage and epithelial cell-derived eotaxin-1 plays a critical role in the regulation of eosinophil recruitment in colonic eosinophilic disease such as pediatric UC and provides a basis for targeting the eosinophil/eotaxin-1 axis in UC.
Conflict of interest statement
The authors have no financial conflict of interest.
Figures


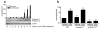


References
-
- Bouma G, Strober W. The immunological and genetic basis of inflammatory bowel disease. Nat Rev Immunol. 2003;3:521–533. - PubMed
-
- Walsh RE, Gaginella TS. The eosinophil in inflammatory bowel disease. Scand J Gastrorenterol. 1991;26:1217–1224. - PubMed
-
- Desreumaux P, Nutten S, Colombel JF. Activated eosinophils in inflammatory bowel disease: do they matter? Am J Gastroenterol. 1999;94:3396 –3398. - PubMed
-
- Nishitani H, Okabayashi M, Satomi M, Shimoyama T, Dohi Y. Infiltration of peroxidase-producing eosinophils into the lamina propria of patients with ulcerative colitis. J Gastroenterol. 1998;33:189 –195. - PubMed
-
- Rothenberg ME, Hogan SP. The eosinophil. Annu Rev Immunol. 2006;24:147–174. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

