P bodies promote stress granule assembly in Saccharomyces cerevisiae
- PMID: 18981231
- PMCID: PMC2575786
- DOI: 10.1083/jcb.200807043
P bodies promote stress granule assembly in Saccharomyces cerevisiae
Abstract
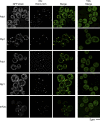
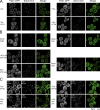
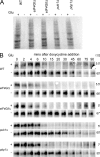
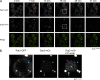
Recent results indicate that nontranslating mRNAs in eukaryotic cells exist in distinct biochemical states that accumulate in P bodies and stress granules, although the nature of interactions between these particles is unknown. We demonstrate in Saccharomyces cerevisiae that RNA granules with similar protein composition and assembly mechanisms as mammalian stress granules form during glucose deprivation. Stress granule assembly is dependent on P-body formation, whereas P-body assembly is independent of stress granule formation. This suggests that stress granules primarily form from mRNPs in preexisting P bodies, which is also supported by the kinetics of P-body and stress granule formation both in yeast and mammalian cells. These observations argue that P bodies are important sites for decisions of mRNA fate and that stress granules, at least in yeast, primarily represent pools of mRNAs stalled in the process of reentry into translation from P bodies.
Figures




References
-
- Abramoff, M.D., P.J. Magelhaes, and S.J. Ram. 2004. Image processing with ImageJ. Biophotonics Intl. 11:36–42.
-
- Anderson, P., and N. Kedersha. 2008. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33:141–150. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

