DNA double-strand break processing: the beginning of the end
- PMID: 18981468
- PMCID: PMC2751443
- DOI: 10.1101/gad.1742408
DNA double-strand break processing: the beginning of the end
Abstract
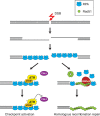
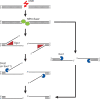
Nucleolytic processing of DNA double-strand breaks (DSBs) generates 3' ssDNA tails that are essential for the assembly of DNA damage checkpoint signaling and DNA repair protein complexes. Genetic studies have provided evidence that multiple nuclease activities are involved in DSB end resection. Three recent studies, including work by Jackson and colleagues (pp. 2767- 2772) in the October 15, 2008, issue of Genes & Development, have begun to shed some light on the intricacy of this process.
Figures
Comment on
-
DNA helicases Sgs1 and BLM promote DNA double-strand break resection.Genes Dev. 2008 Oct 15;22(20):2767-72. doi: 10.1101/gad.503108. Genes Dev. 2008. PMID: 18923075 Free PMC article.
References
-
- Anderson D.G., Kowalczykowski S.C. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a χ-regulated manner. Cell. 1997;90:77–86. - PubMed
-
- Bae S.H., Bae K.H., Kim J.A., Seo Y.S. RPA governs endonucleases switching during processing of Okazaki fragments in eukaryotes. Nature. 2001;412:456–461. - PubMed
-
- Budd M.E., Choe W., Campbell J.L. The nuclease activity of the yeast DNA2 protein, which is related to the RecB-like nucleases, is essential in vivo. J. Biol. Chem. 2000;275:16518–16529. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
