PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale
- PMID: 18981473
- PMCID: PMC2577797
- DOI: 10.1101/gad.1709008
PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale
Abstract
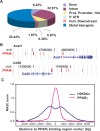
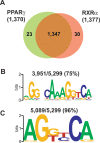
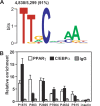
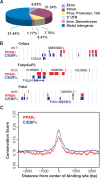
Peroxisome proliferator-activated receptor gamma(PPARgamma), a nuclear receptor and the target of anti-diabetic thiazolinedione drugs, is known as the master regulator of adipocyte biology. Although it regulates hundreds of adipocyte genes, PPARgamma binding to endogenous genes has rarely been demonstrated. Here, utilizing chromatin immunoprecipitation (ChIP) coupled with whole genome tiling arrays, we identified 5299 genomic regions of PPARgamma binding in mouse 3T3-L1 adipocytes. The consensus PPARgamma/RXRalpha "DR-1"-binding motif was found at most of the sites, and ChIP for RXRalpha showed colocalization at nearly all locations tested. Bioinformatics analysis also revealed CCAAT/enhancer-binding protein (C/EBP)-binding motifs in the vicinity of most PPARgamma-binding sites, and genome-wide analysis of C/EBPalpha binding demonstrated that it localized to 3350 of the locations bound by PPARgamma. Importantly, most genes induced in adipogenesis were bound by both PPARgamma and C/EBPalpha, while very few were PPARgamma-specific. C/EBPbeta also plays a role at many of these genes, such that both C/EBPalpha and beta are required along with PPARgamma for robust adipocyte-specific gene expression. Thus, PPARgamma and C/EBP factors cooperatively orchestrate adipocyte biology by adjacent binding on an unanticipated scale.
Figures

References
-
- Anderson P.D., Mehta N.N., Wolfe M.L., Hinkle C.C., Pruscino L., Comiskey L.L., Tabita-Martinez J., Sellers K.F., Rickels M.R., Ahima R.S., et al. Innate immunity modulates adipokines in humans. J. Clin. Endocrinol. Metab. 2007;92:2272–2279. - PubMed
-
- Cao Z., Umek R.M., McKnight S.L. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes & Dev. 1991;5:1538–1552. - PubMed
-
- Carroll J.S., Liu X.S., Brodsky A.S., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. - PubMed
-
- Carroll J.S., Meyer C.A., Song J., Li W., Geistlinger T.R., Eeckhoute J., Brodsky A.S., Keeton E.K., Fertuck K.C., Hall G.F., et al. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 2006;38:1289–1297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
