Emerging roles for centromeres in meiosis I chromosome segregation
- PMID: 18981989
- PMCID: PMC2708604
- DOI: 10.1038/nrg2454
Emerging roles for centromeres in meiosis I chromosome segregation
Abstract
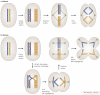
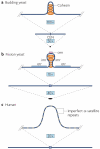
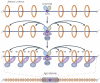
Centromeres are an essential and conserved feature of eukaryotic chromosomes, yet recent research indicates that we are just beginning to understand the numerous roles that centromeres have in chromosome segregation. During meiosis I, in particular, centromeres seem to function in many processes in addition to their canonical role in assembling kinetochores, the sites of microtubule attachment. Here we summarize recent advances that place centromeres at the centre of meiosis I, and discuss how these studies affect a variety of basic research fields and thus hold promise for increasing our understanding of human reproductive defects and disease states.
Figures
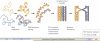

References
-
- Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet. 2001;35:673–745. - PubMed
-
- Lee B, Amon A. Meiosis: how to create a specialized cell cycle. Curr Opin Cell Biol. 2001;13:770–7. - PubMed
-
- Marston AL, Amon A. Meiosis: cell-cycle controls shuffle and deal. Nat Rev Mol Cell Biol. 2004;5:983–97. - PubMed
-
- Sharp L. Introduction to Cytology. McGraw-Hill; New York and London: 1934.
-
- Fitzgerald-Hayes M, Clarke L, Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–44. - PubMed
-
The authors determine a 25 bp sequence from budding yeast to be sufficient for plasmid segregation. This sequence is conserved between chromosomes. This study represents the first identification of a discrete centromere sequence.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

