Specificity and reactivity in menaquinone biosynthesis: the structure of Escherichia coli MenD (2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexadiene-1-carboxylate synthase)
- PMID: 18983854
- PMCID: PMC3656419
- DOI: 10.1016/j.jmb.2008.10.048
Specificity and reactivity in menaquinone biosynthesis: the structure of Escherichia coli MenD (2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexadiene-1-carboxylate synthase)
Abstract
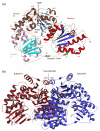
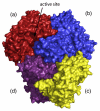
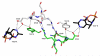
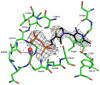
The thiamine diphosphate (ThDP) and metal-ion-dependent enzyme 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexadiene-1-carboxylate synthase, or MenD, catalyze the Stetter-like conjugate addition of alpha-ketoglutarate with isochorismate to release 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexadiene-1-carboxylate and carbon dioxide. This reaction represents the first committed step for biosynthesis of menaquinone, or vitamin K2, a key cofactor for electron transport in bacteria and a metabolite for posttranslational modification of proteins in mammals. The medium-resolution structure of MenD from Escherichia coli (EcMenD) in complex with its cofactor and Mn2+ has been determined in two related hexagonal crystal forms. The subunit displays the typical three-domain structure observed for ThDP-dependent enzymes in which two of the domains bind and force the cofactor into a configuration that supports formation of a reactive ylide. The structures reveal a stable dimer-of-dimers association in agreement with gel filtration and analytical ultracentrifugation studies and confirm the classification of MenD in the pyruvate oxidase family of ThDP-dependent enzymes. The active site, created by contributions from a pair of subunits, is highly basic with a pronounced hydrophobic patch. These features, formed by highly conserved amino acids, match well to the chemical properties of the substrates. A model of the covalent intermediate formed after reaction with the first substrate alpha-ketoglutarate and with the second substrate isochorismate positioned to accept nucleophilic attack has been prepared. This, in addition to structural and sequence comparisons with putative MenD orthologues, provides insight into the specificity and reactivity of MenD and allows a two-stage reaction mechanism to be proposed.
Figures




Similar articles
-
Structure and reactivity of Bacillus subtilis MenD catalyzing the first committed step in menaquinone biosynthesis.J Mol Biol. 2010 Aug 13;401(2):253-64. doi: 10.1016/j.jmb.2010.06.025. Epub 2010 Jun 18. J Mol Biol. 2010. PMID: 20600129 Free PMC article.
-
Structural insights of the MenD from Escherichia coli reveal ThDP affinity.Biochem Biophys Res Commun. 2009 Mar 20;380(4):797-801. doi: 10.1016/j.bbrc.2009.01.168. Epub 2009 Feb 4. Biochem Biophys Res Commun. 2009. PMID: 19338755
-
Two active site arginines are critical determinants of substrate binding and catalysis in MenD: a thiamine-dependent enzyme in menaquinone biosynthesis.Biochem J. 2018 Nov 30;475(22):3651-3667. doi: 10.1042/BCJ20180548. Biochem J. 2018. PMID: 30341164
-
Steady-state kinetics and molecular evolution of Escherichia coli MenD [(1R,6R)-2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase], an anomalous thiamin diphosphate-dependent decarboxylase-carboligase.Biochemistry. 2003 Nov 25;42(46):13496-504. doi: 10.1021/bi035224j. Biochemistry. 2003. PMID: 14621995
-
Using site-saturation mutagenesis to explore mechanism and substrate specificity in thiamin diphosphate-dependent enzymes.FEBS J. 2013 Dec;280(24):6395-411. doi: 10.1111/febs.12459. Epub 2013 Aug 23. FEBS J. 2013. PMID: 23895593 Review.
Cited by
-
Structures of Listeria monocytogenes MenD in ThDP-bound and in-crystallo captured intermediate I-bound forms.Acta Crystallogr F Struct Biol Commun. 2025 Aug 1;81(Pt 8):348-357. doi: 10.1107/S2053230X25006181. Epub 2025 Jul 17. Acta Crystallogr F Struct Biol Commun. 2025. PMID: 40673487 Free PMC article.
-
The Enzymology of Organic Transformations: A Survey of Name Reactions in Biological Systems.Angew Chem Int Ed Engl. 2017 Mar 20;56(13):3446-3489. doi: 10.1002/anie.201603291. Epub 2017 Feb 14. Angew Chem Int Ed Engl. 2017. PMID: 27505692 Free PMC article. Review.
-
Allosteric regulation of menaquinone (vitamin K2) biosynthesis in the human pathogen Mycobacterium tuberculosis.J Biol Chem. 2020 Mar 20;295(12):3759-3770. doi: 10.1074/jbc.RA119.012158. Epub 2020 Feb 6. J Biol Chem. 2020. PMID: 32029475 Free PMC article.
-
Effects of Alkali Stress on the Growth and Menaquinone-7 Metabolism of Bacillus subtilis natto.Front Microbiol. 2022 Apr 28;13:899802. doi: 10.3389/fmicb.2022.899802. eCollection 2022. Front Microbiol. 2022. PMID: 35572665 Free PMC article.
-
Structure of Staphylococcus aureus adenylosuccinate lyase (PurB) and assessment of its potential as a target for structure-based inhibitor discovery.Acta Crystallogr D Biol Crystallogr. 2010 Aug;66(Pt 8):881-8. doi: 10.1107/S0907444910020081. Epub 2010 Jul 9. Acta Crystallogr D Biol Crystallogr. 2010. PMID: 20693687 Free PMC article.
References
-
- Meganathan R. Biosynthesis of the isoprenoid quinones menaquinone (vitamin K2) and ubiquinone (coenzyme Q) In: Neidhardt FC, Curtiss R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. American Society for Microbiology; Washington, DC: 1996. pp. 642–656.
-
- Meganathan R. Biosynthesis of menaquinone (vitamin K2) and ubiquinone (coenzyme Q): a perspective on enzymatic mechanisms. Vitam. Horm. 2001;61:173–218. - PubMed
-
- Soballe B, Poole RK. Microbial ubiquinones: multiple roles in respiration, gene regulation and oxidative stress management. Microbiology. 1999;145:1817–1830. - PubMed
-
- Bekker M, Kramer G, Hartog AF, Wagner MJ, de Koster CG, Hellingwerf KJ, de Mattos MJ. Changes in the redox state and composition of the quinone pool of Escherichia coli during aerobic batch-culture growth. Microbiology. 2007;153:1974–1980. - PubMed
-
- Bandyopadhyay PK. Vitamin K-dependent gamma-glutamylcarboxylation: an ancient posttranslational modification. Vitam. Horm. 2008;78:157–184. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

