Kinetic analysis of correct nucleotide insertion by a Y-family DNA polymerase reveals conformational changes both prior to and following phosphodiester bond formation as detected by tryptophan fluorescence
- PMID: 18984592
- PMCID: PMC2605995
- DOI: 10.1074/jbc.M806785200
Kinetic analysis of correct nucleotide insertion by a Y-family DNA polymerase reveals conformational changes both prior to and following phosphodiester bond formation as detected by tryptophan fluorescence
Abstract
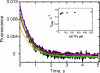
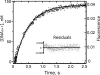
The Sulfolobus solfataricus Y-family DNA polymerase Dpo4 is a model for translesion replication and has been used in the analysis of individual steps involved in catalysis. The role of conformational changes has not been clear. Introduction of Trp residues into the Trp-devoid wild-type protein provided fluorescence probes of these events, particularly in the case of mutants T239W and N188W. With both mutants, a rapid increase in Trp fluorescence was observed only in the case of normal base pairing (G:C), was saturable with respect to dCTP concentration, and occurred in the absence of phosphodiester bond formation. A subsequent decrease in the Trp fluorescence occurred when phosphodiester bond formation was permitted, and these rates were independent of the dCTP concentration. This step is relatively slow and is attributed to a conformational relaxation step occurring after pyrophosphate release, which was measured and shown to be fast in a separate experiment. The measured rate of release of DNA from Dpo4 was rapid and is not rate-limiting. Overall, the measurements provide a kinetic scheme for Dpo4 different than generally accepted for replicative polymerases or proposed for Dpo4 and other Y-family polymerases: the initial enzyme.DNA.dNTP complex undergoes a rapid (18 s(-1)), reversible (21 s(-1)) conformational change, followed by relatively rapid phosphodiester bond formation (11 s(-1)) and then fast release of pyrophosphate, followed by a rate-limiting relaxation of the active conformation (2 s(-1)) and then rapid DNA release, yielding an overall steady-state kcat of <1 s(-1).
Figures








References
-
- Kunkel, T. A. (2004) J. Biol. Chem. 279 16895-16898 - PubMed
-
- Prakash, S., Johnson, R. E., and Prakash, L. (2004) Annu. Rev. Biochem. 74 317-353 - PubMed
-
- Kool, E. T. (2002) Annu. Rev. Biochem. 71 191-219 - PubMed
-
- Lehmann, A. R., Niimi, A., Ogi, T., Brown, S., Sabbioneda, S., Wing, J. F., Kannouche, P. L., and Green, C. M. (2007) DNA Repair (Amst.) 6 891-899 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

